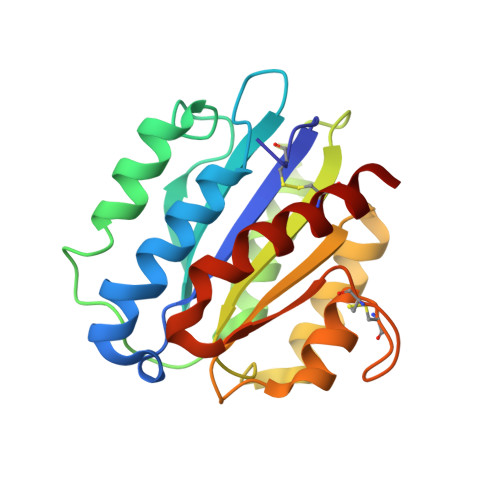
Structure of Plasmodium falciparum TRAP (thrombospondin-related anonymous protein) A domain highlights distinct features in apicomplexan von Willebrand factor A homologues.
Pihlajamaa, T., Kajander, T., Knuuti, J., Horkka, K., Sharma, A., Permi, P.(2013) Biochem J 450: 469-476
- PubMed: 23317521
- DOI: https://doi.org/10.1042/BJ20121058
- Primary Citation of Related Structures:
4F1J, 4F1K - PubMed Abstract:
TRAP (thrombospondin-related anonymous protein), localized in the micronemes and on the surface of sporozoites of the notorious malaria parasite Plasmodium, is a key molecule upon infection of mammalian host hepatocytes and invasion of mosquito salivary glands. TRAP contains two adhesive domains responsible for host cell recognition and invasion, and is known to be essential for infectivity. In the present paper, we report high-resolution crystal structures of the A domain of Plasmodium falciparum TRAP with and without bound Mg2+. The structure reveals a vWA (von Willebrand factor A)-like fold and a functional MIDAS (metal-ion-dependent adhesion site), as well as a potential heparan sulfate-binding site. Site-directed mutagenesis and cell-attachment assays were used to investigate the functional roles of the surface epitopes discovered. The reported structures are the first determined for a complete vWA domain of parasitic origin, highlighting unique features among homologous domains from other proteins characterized hitherto. Some of these are conserved among Plasmodiae exclusively, whereas others may be common to apicomplexan organisms in general.
- Program in Structural Biology and Biophysics, Institute of Biotechnology, P.O. Box 65, FI-00014 University of Helsinki, Helsinki, Finland.
Organizational Affiliation: