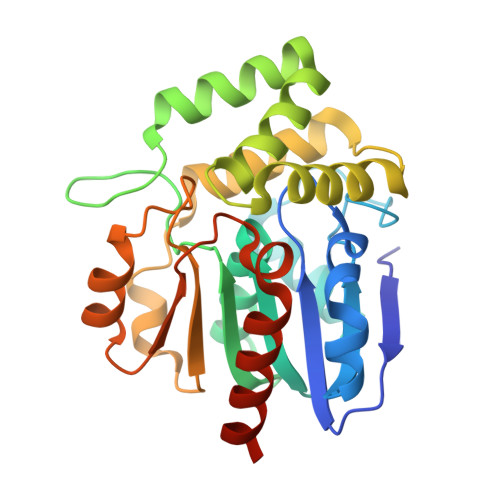
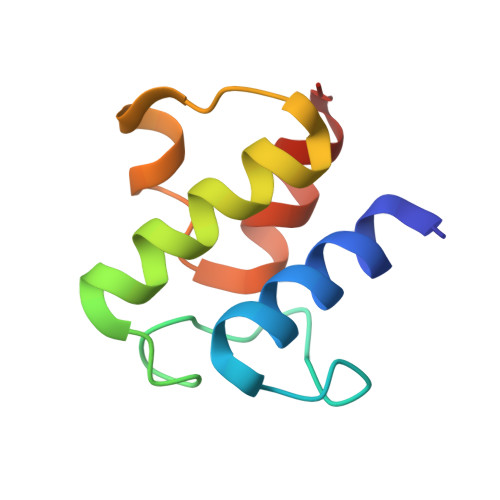
Structure of the enzyme-acyl carrier protein (ACP) substrate gatekeeper complex required for biotin synthesis.
Agarwal, V., Lin, S., Lukk, T., Nair, S.K., Cronan, J.E.(2012) Proc Natl Acad Sci U S A 109: 17406-17411
- PubMed: 23045647
- DOI: https://doi.org/10.1073/pnas.1207028109
- Primary Citation of Related Structures:
4ETW - PubMed Abstract:
Although the pimeloyl moiety was long known to be a biotin precursor, the mechanism of assembly of this C7 α,ω-dicarboxylic acid was only recently elucidated. In Escherichia coli, pimelate is made by bypassing the strict specificity of the fatty acid synthetic pathway. BioC methylates the free carboxyl of a malonyl thioester, which replaces the usual acetyl thioester primer. This atypical primer is transformed to pimeloyl-acyl carrier protein (ACP) methyl ester by two cycles of fatty acid synthesis. The question is, what stops this product from undergoing further elongation? Although BioH readily cleaves this product in vitro, the enzyme is nonspecific, which made assignment of its physiological substrate problematical, especially because another enzyme, BioF, could also perform this gatekeeping function. We report the 2.05-Å resolution cocrystal structure of a complex of BioH with pimeloyl-ACP methyl ester and use the structure to demonstrate that BioH is the gatekeeper and its physiological substrate is pimeloyl-ACP methyl ester.
- Center for Biophysics and Computational Biology, Institute for Genomic Biology, Department of Microbiology and Biochemistry, University of Illinois, Urbana, IL 61801, USA.
Organizational Affiliation: