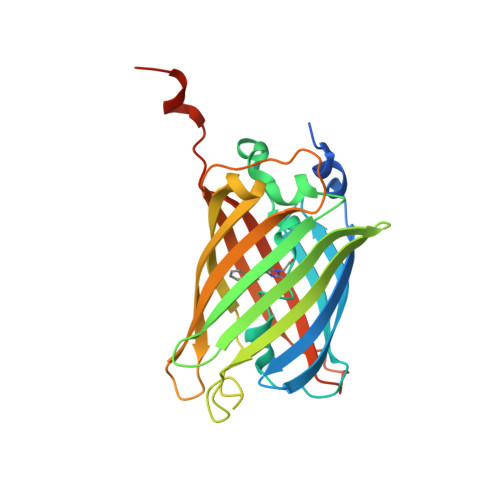
The 1.6 A resolution structure of a FRET-optimized Cerulean fluorescent protein.
Watkins, J.L., Kim, H., Markwardt, M.L., Chen, L., Fromme, R., Rizzo, M.A., Wachter, R.M.(2013) Acta Crystallogr D Biol Crystallogr 69: 767-773
- PubMed: 23633585
- DOI: https://doi.org/10.1107/S0907444913001546
- Primary Citation of Related Structures:
4EN1 - PubMed Abstract:
Genetically encoded cyan fluorescent proteins (CFPs) bearing a tryptophan-derived chromophore are commonly used as energy-donor probes in Förster resonance energy transfer (FRET) experiments useful in live cell-imaging applications. In recent years, significant effort has been expended on eliminating the structural and excited-state heterogeneity of these proteins, which has been linked to undesirable photophysical properties. Recently, mCerulean3, a descendant of enhanced CFP, was introduced as an optimized FRET donor protein with a superior quantum yield of 0.87. Here, the 1.6 Å resolution X-ray structure of mCerulean3 is reported. The chromophore is shown to adopt a planar trans configuration at low pH values, indicating that the acid-induced isomerization of Cerulean has been eliminated. β-Strand 7 appears to be well ordered in a single conformation, indicating a loss of conformational heterogeneity in the vicinity of the chromophore. Although the side chains of Ile146 and Leu167 appear to exist in two rotamer states, they are found to be well packed against the indole group of the chromophore. The Ser65 reversion mutation allows improved side-chain packing of Leu220. A structural comparison with mTurquoise2 is presented and additional engineering strategies are discussed.
- Department of Chemistry and Biochemistry, Arizona State University, Tempe, AZ 85287-1604, USA.
Organizational Affiliation: