Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors.
Ashwell, M.A., Lapierre, J.M., Brassard, C., Bresciano, K., Bull, C., Cornell-Kennon, S., Eathiraj, S., France, D.S., Hall, T., Hill, J., Kelleher, E., Khanapurkar, S., Kizer, D., Koerner, S., Link, J., Liu, Y., Makhija, S., Moussa, M., Namdev, N., Nguyen, K., Nicewonger, R., Palma, R., Szwaya, J., Tandon, M., Uppalapati, U., Vensel, D., Volak, L.P., Volckova, E., Westlund, N., Wu, H., Yang, R.Y., Chan, T.C.(2012) J Med Chem 55: 5291-5310
- PubMed: 22533986
- DOI: https://doi.org/10.1021/jm300276x
- Primary Citation of Related Structures:
4EJN - PubMed Abstract:
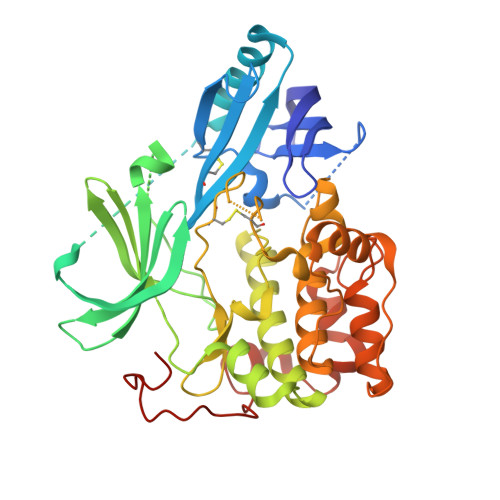
This paper describes the implementation of a biochemical and biophysical screening strategy to identify and optimize small molecule Akt1 inhibitors that act through a mechanism distinct from that observed for kinase domain ATP-competitive inhibitors. With the aid of an unphosphorylated Akt1 cocrystal structure of 12j solved at 2.25 Å, it was possible to confirm that as a consequence of binding these novel inhibitors, the ATP binding cleft contained a number of hydrophobic residues that occlude ATP binding as expected. These Akt inhibitors potently inhibit intracellular Akt activation and its downstream target (PRAS40) in vitro. In vivo pharmacodynamic and pharmacokinetic studies with two examples, 12e and 12j, showed the series to be similarly effective at inhibiting the activation of Akt and an additional downstream effector (p70S6) following oral dosing in mice.
- ArQule Inc., 19 Presidential Way, Woburn, Massachusetts 01801, United States.
Organizational Affiliation: