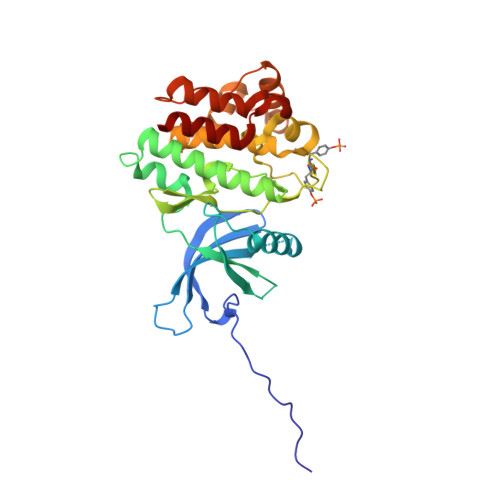
Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
Zak, M., Mendonca, R., Balazs, M., Barrett, K., Bergeron, P., Blair, W.S., Chang, C., Deshmukh, G., Devoss, J., Dragovich, P.S., Eigenbrot, C., Ghilardi, N., Gibbons, P., Gradl, S., Hamman, C., Hanan, E.J., Harstad, E., Hewitt, P.R., Hurley, C.A., Jin, T., Johnson, A., Johnson, T., Kenny, J.R., Koehler, M.F., Bir Kohli, P., Kulagowski, J.J., Labadie, S., Liao, J., Liimatta, M., Lin, Z., Lupardus, P.J., Maxey, R.J., Murray, J.M., Pulk, R., Rodriguez, M., Savage, S., Shia, S., Steffek, M., Ubhayakar, S., Ultsch, M., van Abbema, A., Ward, S.I., Xiao, L., Xiao, Y.(2012) J Med Chem 55: 6176-6193
- PubMed: 22698084
- DOI: https://doi.org/10.1021/jm300628c
- Primary Citation of Related Structures:
4EHZ, 4EI4, 4F08, 4F09 - PubMed Abstract:
Herein we report the discovery of the C-2 methyl substituted imidazopyrrolopyridine series and its optimization to provide potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2. The C-2 methyl substituted inhibitor 4 exhibited not only improved JAK1 potency relative to unsubstituted compound 3 but also notable JAK1 vs JAK2 selectivity (20-fold and >33-fold in biochemical and cell-based assays, respectively). Features of the X-ray structures of 4 in complex with both JAK1 and JAK2 are delineated. Efforts to improve the in vitro and in vivo ADME properties of 4 while maintaining JAK1 selectivity are described, culminating in the discovery of a highly optimized and balanced inhibitor (20). Details of the biological characterization of 20 are disclosed including JAK1 vs JAK2 selectivity levels, preclinical in vivo PK profiles, performance in an in vivo JAK1-mediated PK/PD model, and attributes of an X-ray structure in complex with JAK1.
- Department of Discovery Chemistry, Genentech, Inc, South San Francisco, CA 94080, USA. mzak@gene.com
Organizational Affiliation: