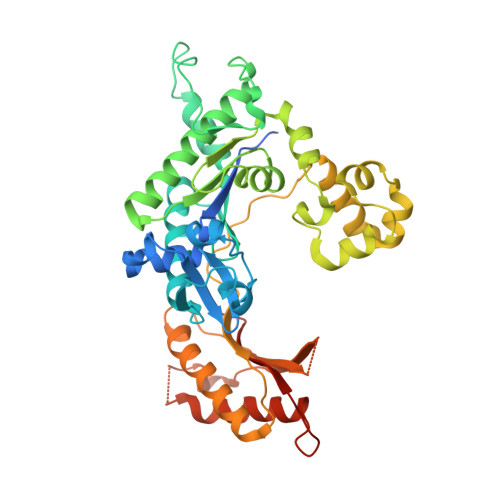
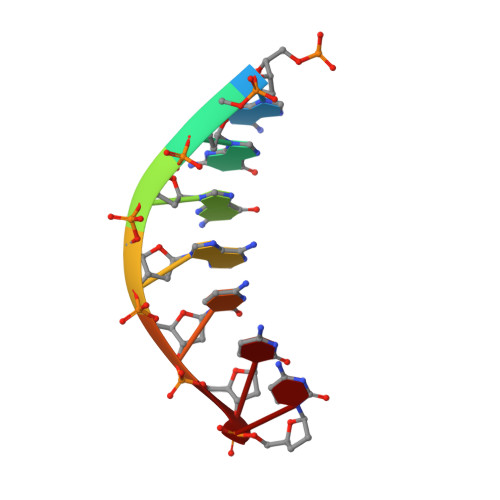
A Nucleotide-Analogue-Induced Gain of Function Corrects the Error-Prone Nature of Human DNA Polymerase iota.
Ketkar, A., Zafar, M.K., Banerjee, S., Marquez, V.E., Egli, M., Eoff, R.L.(2012) J Am Chem Soc 134: 10698-10705
- PubMed: 22632140
- DOI: https://doi.org/10.1021/ja304176q
- Primary Citation of Related Structures:
4EBC, 4EBD, 4EBE - PubMed Abstract:
Y-family DNA polymerases participate in replication stress and DNA damage tolerance mechanisms. The properties that allow these enzymes to copy past bulky adducts or distorted template DNA can result in a greater propensity for them to make mistakes. Of the four human Y-family members, human DNA polymerase iota (hpol ι) is the most error-prone. In the current study, we elucidate the molecular basis for improving the fidelity of hpol ι through use of the fixed-conformation nucleotide North-methanocarba-2'-deoxyadenosine triphosphate (N-MC-dATP). Three crystal structures were solved of hpol ι in complex with DNA containing a template 2'-deoxythymidine (dT) paired with an incoming dNTP or modified nucleotide triphosphate. The ternary complex of hpol ι inserting N-MC-dATP opposite dT reveals that the adenine ring is stabilized in the anti orientation about the pseudo-glycosyl torsion angle, which mimics precisely the mutagenic arrangement of dGTP:dT normally preferred by hpol ι. The stabilized anti conformation occurs without notable contacts from the protein but likely results from constraints imposed by the bicyclo[3.1.0]hexane scaffold of the modified nucleotide. Unmodified dATP and South-MC-dATP each adopt syn glycosyl orientations to form Hoogsteen base pairs with dT. The Hoogsteen orientation exhibits weaker base-stacking interactions and is less catalytically favorable than anti N-MC-dATP. Thus, N-MC-dATP corrects the error-prone nature of hpol ι by preventing the Hoogsteen base-pairing mode normally observed for hpol ι-catalyzed insertion of dATP opposite dT. These results provide a previously unrecognized means of altering the efficiency and the fidelity of a human translesion DNA polymerase.
- Department of Biochemistry and Molecular Biology, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205-7199, USA.
Organizational Affiliation: