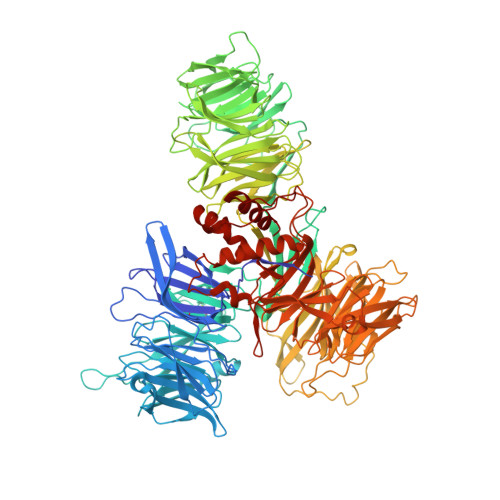
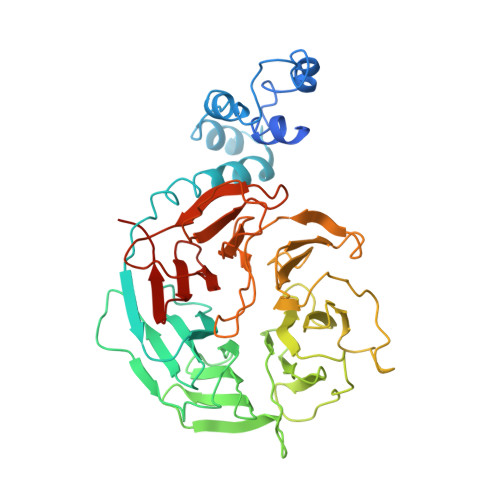
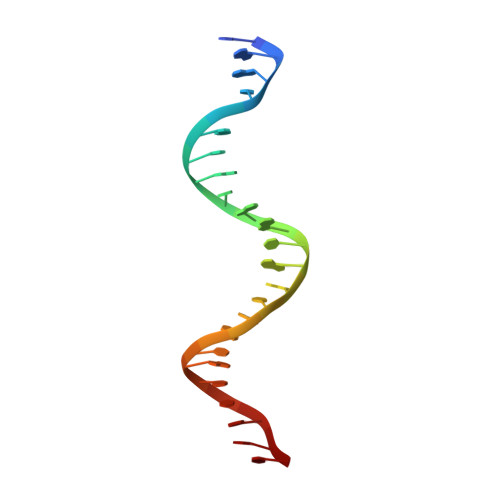
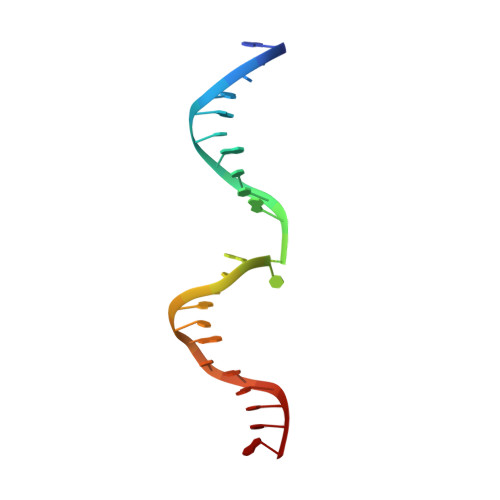
Damaged DNA induced UV-damaged DNA-binding protein (UV-DDB) dimerization and its roles in chromatinized DNA repair.
Yeh, J.I., Levine, A.S., Du, S., Chinte, U., Ghodke, H., Wang, H., Shi, H., Hsieh, C.L., Conway, J.F., Van Houten, B., Rapic-Otrin, V.(2012) Proc Natl Acad Sci U S A 109: E2737-E2746
- PubMed: 22822215
- DOI: https://doi.org/10.1073/pnas.1110067109
- Primary Citation of Related Structures:
4E54, 4E5Z - PubMed Abstract:
UV light-induced photoproducts are recognized and removed by the nucleotide-excision repair (NER) pathway. In humans, the UV-damaged DNA-binding protein (UV-DDB) is part of a ubiquitin E3 ligase complex (DDB1-CUL4A(DDB2)) that initiates NER by recognizing damaged chromatin with concomitant ubiquitination of core histones at the lesion. We report the X-ray crystal structure of the human UV-DDB in a complex with damaged DNA and show that the N-terminal domain of DDB2 makes critical contacts with two molecules of DNA, driving N-terminal-domain folding and promoting UV-DDB dimerization. The functional significance of the dimeric UV-DDB [(DDB1-DDB2)(2)], in a complex with damaged DNA, is validated by electron microscopy, atomic force microscopy, solution biophysical, and functional analyses. We propose that the binding of UV-damaged DNA results in conformational changes in the N-terminal domain of DDB2, inducing helical folding in the context of the bound DNA and inducing dimerization as a function of nucleotide binding. The temporal and spatial interplay between domain ordering and dimerization provides an elegant molecular rationale for the unprecedented binding affinities and selectivities exhibited by UV-DDB for UV-damaged DNA. Modeling the DDB1-CUL4A(DDB2) complex according to the dimeric UV-DDB-AP24 architecture results in a mechanistically consistent alignment of the E3 ligase bound to a nucleosome harboring damaged DNA. Our findings provide unique structural and conformational insights into the molecular architecture of the DDB1-CUL4A(DDB2) E3 ligase, with significant implications for the regulation and overall organization of the proteins responsible for initiation of NER in the context of chromatin and for the consequent maintenance of genomic integrity.
- Department of Structural Biology, University of Pittsburgh School of Medicine, Pittsburgh, PA 15260, USA. jiyeh@pitt.edu
Organizational Affiliation: