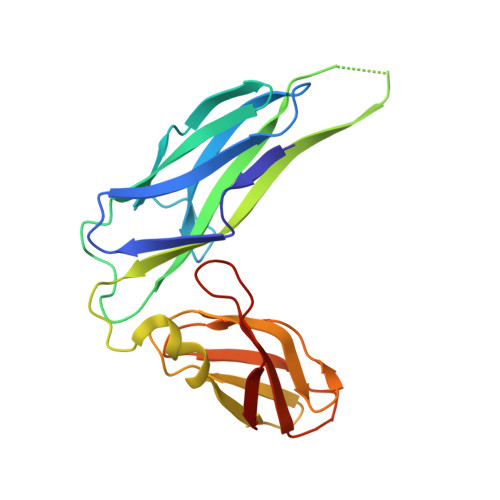
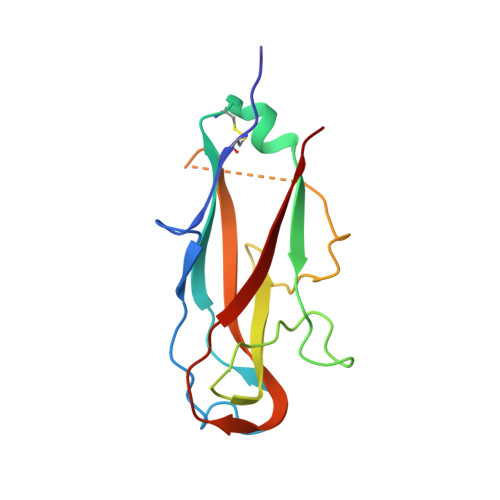
Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Crespo, M.D., Puorger, C., Scharer, M.A., Eidam, O., Grutter, M.G., Capitani, G., Glockshuber, R.(2012) Nat Chem Biol 8: 707-713
- PubMed: 22772153
- DOI: https://doi.org/10.1038/nchembio.1019
- Primary Citation of Related Structures:
3SQB, 4DWH - PubMed Abstract:
Type 1 pili from uropathogenic Escherichia coli are filamentous, noncovalent protein complexes mediating bacterial adhesion to the host tissue. All structural pilus subunits are homologous proteins sharing an invariant disulfide bridge. Here we show that disulfide bond formation in the unfolded subunits, catalyzed by the periplasmic oxidoreductase DsbA, is required for subunit recognition by the assembly chaperone FimC and for FimC-catalyzed subunit folding. FimC thus guarantees quantitative disulfide bond formation in each of the up to 3,000 subunits of the pilus. The X-ray structure of the complex between FimC and the main pilus subunit FimA and the kinetics of FimC-catalyzed FimA folding indicate that FimC accelerates folding of pilus subunits by lowering their topological complexity. The kinetic data, together with the measured in vivo concentrations of DsbA and FimC, predict an in vivo half-life of 2 s for oxidative folding of FimA in the periplasm.
- Department of Biology, Institute of Molecular Biology and Biophysics, Swiss Federal Institute of Technology Zurich, Zurich, Switzerland.
Organizational Affiliation: