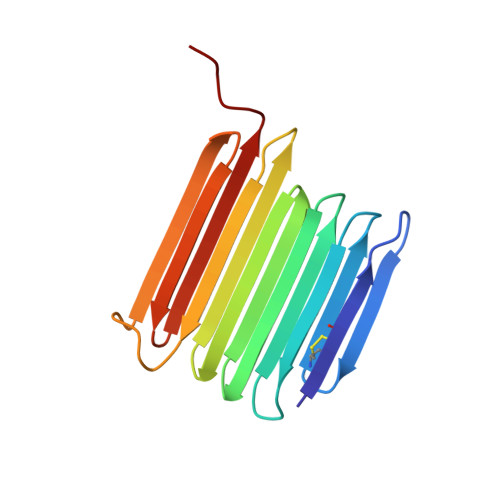
Crystal structure of an insect antifreeze protein and its implications for ice binding.
Hakim, A., Nguyen, J.B., Basu, K., Zhu, D.F., Thakral, D., Davies, P.L., Isaacs, F.J., Modis, Y., Meng, W.(2013) J Biological Chem 288: 12295-12304
- PubMed: 23486477
- DOI: https://doi.org/10.1074/jbc.M113.450973
- Primary Citation of Related Structures:
4DT5 - PubMed Abstract:
Antifreeze proteins (AFPs) help some organisms resist freezing by binding to ice crystals and inhibiting their growth. The molecular basis for how these proteins recognize and bind ice is not well understood. The longhorn beetle Rhagium inquisitor can supercool to below -25 °C, in part by synthesizing the most potent antifreeze protein studied thus far (RiAFP). We report the crystal structure of the 13-kDa RiAFP, determined at 1.21 Å resolution using direct methods. The structure, which contains 1,914 nonhydrogen protein atoms in the asymmetric unit, is the largest determined ab initio without heavy atoms. It reveals a compressed β-solenoid fold in which the top and bottom sheets are held together by a silk-like interdigitation of short side chains. RiAFP is perhaps the most regular structure yet observed. It is a second independently evolved AFP type in beetles. The two beetle AFPs have in common an extremely flat ice-binding surface comprising regular outward-projecting parallel arrays of threonine residues. The more active, wider RiAFP has four (rather than two) of these arrays between which the crystal structure shows the presence of ice-like waters. Molecular dynamics simulations independently reproduce the locations of these ordered crystallographic waters and predict additional waters that together provide an extensive view of the AFP interaction with ice. By matching several planes of hexagonal ice, these waters may help freeze the AFP to the ice surface, thus providing the molecular basis of ice binding.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520, USA.
Organizational Affiliation: