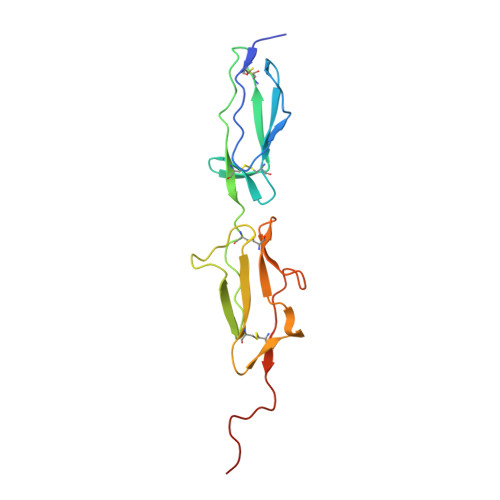
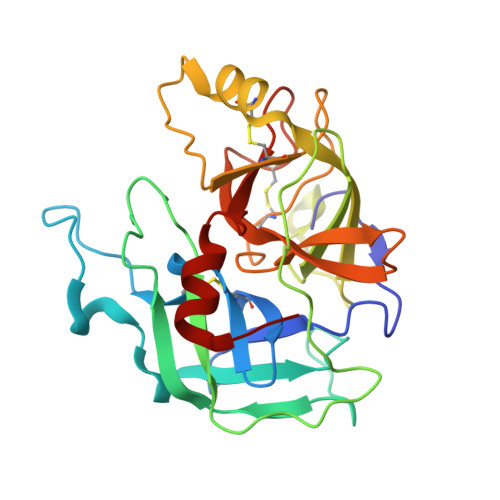
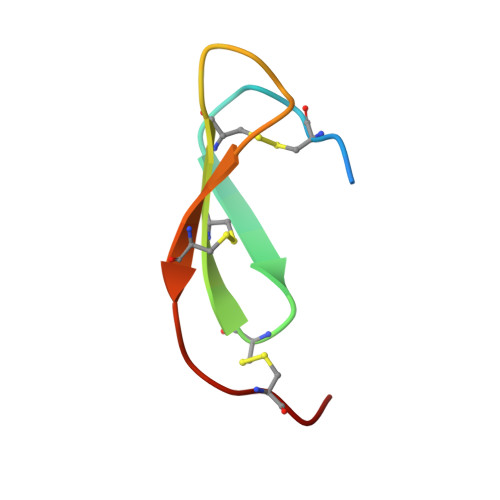
Monospecific Inhibitors Show That Both Mannan-binding Lectin-associated Serine Protease-1 (MASP-1) and -2 Are Essential for Lectin Pathway Activation and Reveal Structural Plasticity of MASP-2.
Heja, D., Harmat, V., Fodor, K., Wilmanns, M., Dobo, J., Kekesi, K.A., Zavodszky, P., Gal, P., Pal, G.(2012) J Biological Chem 287: 20290-20300
- PubMed: 22511776
- DOI: https://doi.org/10.1074/jbc.M112.354332
- Primary Citation of Related Structures:
3TVJ, 4DJZ - PubMed Abstract:
The lectin pathway is an antibody-independent activation route of the complement system. It provides immediate defense against pathogens and altered self-cells, but it also causes severe tissue damage after stroke, heart attack, and other ischemia reperfusion injuries. The pathway is triggered by target binding of pattern recognition molecules leading to the activation of zymogen mannan-binding lectin-associated serine proteases (MASPs). MASP-2 is considered as the autonomous pathway-activator, while MASP-1 is considered as an auxiliary component. We evolved a pair of monospecific MASP inhibitors. In accordance with the key role of MASP-2, the MASP-2 inhibitor completely blocks the lectin pathway activation. Importantly, the MASP-1 inhibitor does the same, demonstrating that MASP-1 is not an auxiliary but an essential pathway component. We report the first Michaelis-like complex structures of MASP-1 and MASP-2 formed with substrate-like inhibitors. The 1.28 Å resolution MASP-2 structure reveals significant plasticity of the protease, suggesting that either an induced fit or a conformational selection mechanism should contribute to the extreme specificity of the enzyme.
- Department of Biochemistry, Eötvös Loránd University, 1/C Pázmány Péter Street, H-1117, Budapest, Hungary.
Organizational Affiliation: