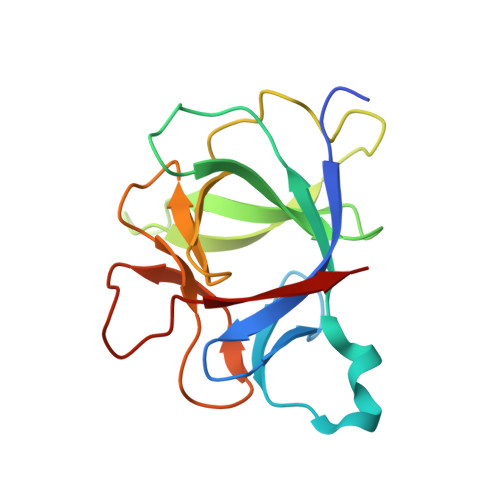
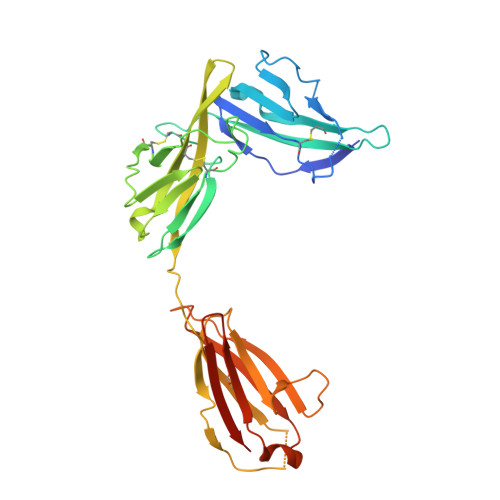
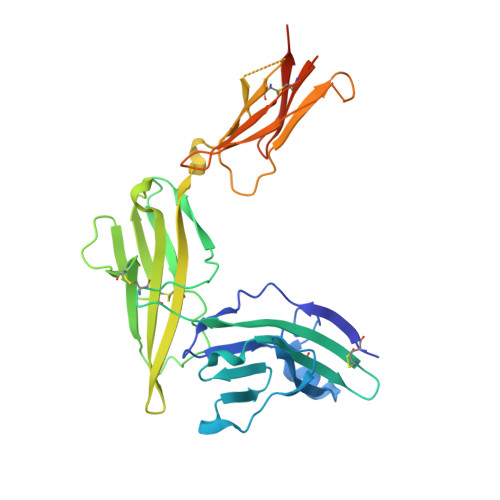
Structure of the activating IL-1 receptor signaling complex.
Thomas, C., Bazan, J.F., Garcia, K.C.(2012) Nat Struct Mol Biol 19: 455-457
- PubMed: 22426547
- DOI: https://doi.org/10.1038/nsmb.2260
- Primary Citation of Related Structures:
4DEP - PubMed Abstract:
Interleukin-1 (IL-1)-family cytokines are mediators of innate and adaptive immunity. They exert proinflammatory effects by binding a primary receptor that recruits a receptor accessory protein to form a signaling-competent heterotrimeric complex. Here we present the crystal structure of IL-1β bound to its primary receptor IL-1RI and its receptor accessory protein IL-1RAcP, providing insight into how IL-1-type cytokines initiate signaling and revealing an evolutionary relationship with the fibroblast growth factor receptor family.
- Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, California, USA.
Organizational Affiliation: