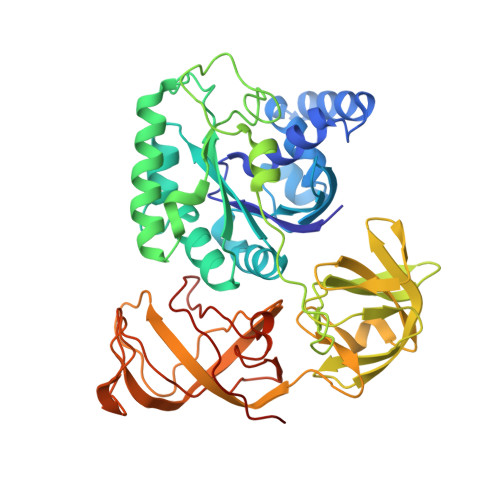
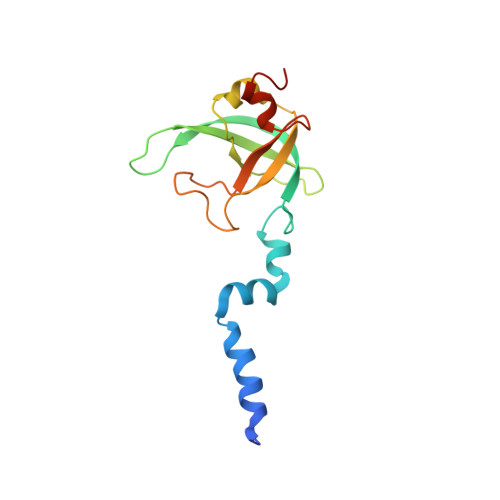
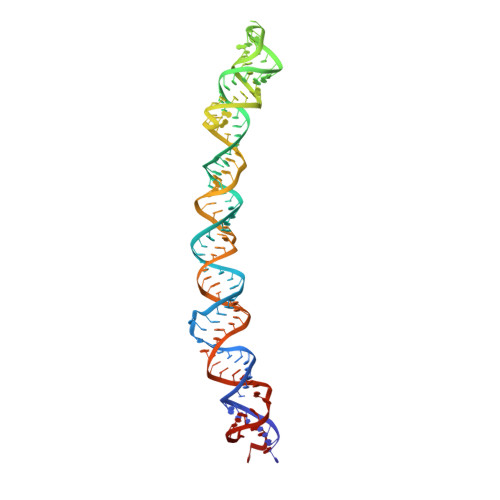
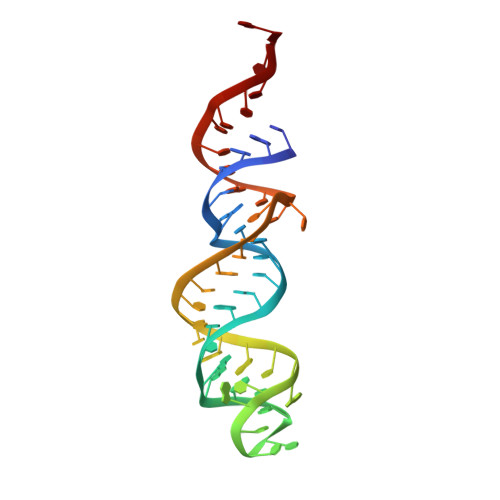
Regulation of the Mammalian Elongation Cycle by Subunit Rolling: A Eukaryotic-Specific Ribosome Rearrangement.
Budkevich, T.V., Giesebrecht, J., Behrmann, E., Loerke, J., Ramrath, D.J., Mielke, T., Ismer, J., Hildebrand, P.W., Tung, C.S., Nierhaus, K.H., Sanbonmatsu, K.Y., Spahn, C.M.(2014) Cell 158: 121
- PubMed: 24995983
- DOI: https://doi.org/10.1016/j.cell.2014.04.044
- Primary Citation of Related Structures:
4CXG, 4CXH, 4UJE - PubMed Abstract:
The extent to which bacterial ribosomes and the significantly larger eukaryotic ribosomes share the same mechanisms of ribosomal elongation is unknown. Here, we present subnanometer resolution cryoelectron microscopy maps of the mammalian 80S ribosome in the posttranslocational state and in complex with the eukaryotic eEF1A⋅Val-tRNA⋅GMPPNP ternary complex, revealing significant differences in the elongation mechanism between bacteria and mammals. Surprisingly, and in contrast to bacterial ribosomes, a rotation of the small subunit around its long axis and orthogonal to the well-known intersubunit rotation distinguishes the posttranslocational state from the classical pretranslocational state ribosome. We term this motion "subunit rolling." Correspondingly, a mammalian decoding complex visualized in substates before and after codon recognition reveals structural distinctions from the bacterial system. These findings suggest how codon recognition leads to GTPase activation in the mammalian system and demonstrate that in mammalia subunit rolling occurs during tRNA selection.
- Institut für Medizinische Physik und Biophysik, Charité-Universitätsmedizin Berlin, Charitéplatz 1, 10117 Berlin, Germany; Max-Planck Institut für Molekulare Genetik, Abteilung Vingron, AG Ribosomen, 14195 Berlin, Ihnestraße 73, Germany; Institute of Molecular Biology and Genetics, Group of Protein Biosynthesis, 03143 Kiev, Ukraine.
Organizational Affiliation: