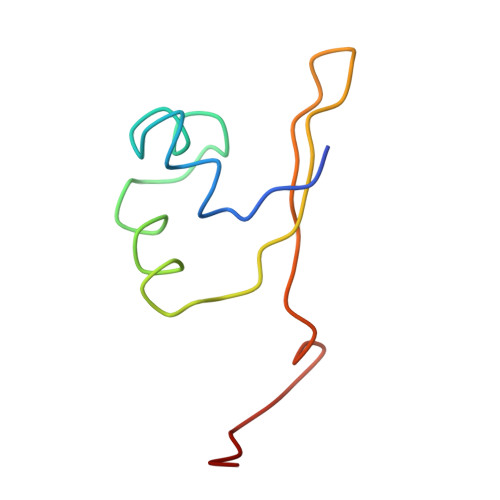
Protein-DNA conformational changes in the crystal structure of a lambda Cro-operator complex.
Brennan, R.G., Roderick, S.L., Takeda, Y., Matthews, B.W.(1990) Proc Natl Acad Sci U S A 87: 8165-8169
- PubMed: 2146682
- DOI: https://doi.org/10.1073/pnas.87.20.8165
- Primary Citation of Related Structures:
4CRO - PubMed Abstract:
The structure of a complex of bacteriophage lambda Cro protein with a 17-base-pair operator has been determined at 3.9-A resolution. Isomorphous derivatives obtained by the synthesis of site-specific iodinated DNA oligomers were of critical importance in solving the structure. The crystal structure contains three independent Cro-operator complexes that have very similar, although not necessarily identical, conformations. In the complex, the protein dimer undergoes a large conformational change relative to the crystal structure of the free protein. One monomer rotates by about 40 degrees relative to the other, this being accomplished primarily by a twisting of the two beta-sheet strands that connect one monomer with the other. In the complex, the DNA is bent by about 40 degrees into the shape of a boomerang but maintains essentially Watson-Crick B-form. In contrast to other known protein-DNA complexes, the DNA is not stacked end-to-end. The structure confirms the general features of the model previously proposed for the interaction of Cro with DNA.
- Department of Physics, University of Oregon, Eugene 97403.
Organizational Affiliation: