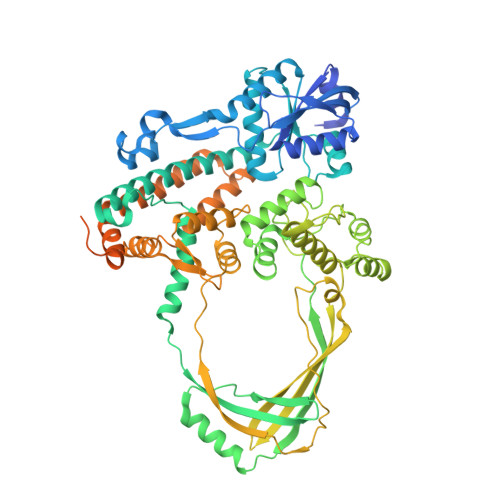
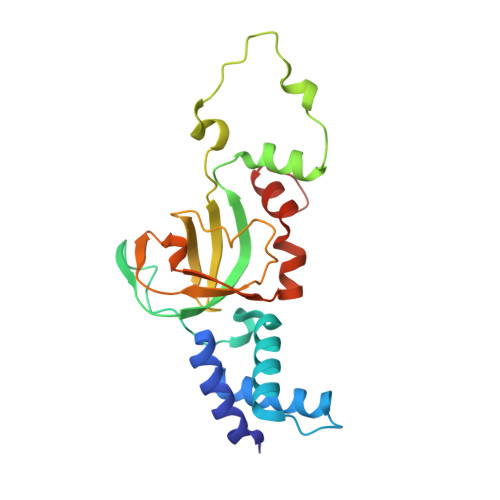
Structural and Mechanistic Insight Into Holliday-Junction Dissolution by Topoisomerase Iiialpha and Rmi1
Bocquet, N., Bizard, A.H., Abdulrahman, W., Larsen, N.B., Faty, M., Cavadini, S., Bunker, R.D., Kowalczykowski, S.C., Cejka, P., Hickson, I.D., Thoma, N.H.(2014) Nat Struct Mol Biol 21: 261
- PubMed: 24509834
- DOI: https://doi.org/10.1038/nsmb.2775
- Primary Citation of Related Structures:
4CGY, 4CHT - PubMed Abstract:
Repair of DNA double-strand breaks via homologous recombination can produce double Holliday junctions (dHJs) that require enzymatic separation. Topoisomerase IIIα (TopIIIα) together with RMI1 disentangles the final hemicatenane intermediate obtained once dHJs have converged. How binding of RMI1 to TopIIIα influences it to behave as a hemicatenane dissolvase, rather than as an enzyme that relaxes DNA topology, is unknown. Here, we present the crystal structure of human TopIIIα complexed to the first oligonucleotide-binding domain (OB fold) of RMI1. TopIII assumes a toroidal type 1A topoisomerase fold. RMI1 attaches to the edge of the gate in TopIIIα through which DNA passes. RMI1 projects a 23-residue loop into the TopIIIα gate, thereby influencing the dynamics of its opening and closing. Our results provide a mechanistic rationale for how RMI1 stabilizes TopIIIα-gate opening to enable dissolution and illustrate how binding partners modulate topoisomerase function.
- Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland.
Organizational Affiliation: