Catalytic features and crystal structure of a tau class glutathione transferase from Glycine max specifically upregulated in response to soybean mosaic virus infections.
Skopelitou, K., Muleta, A.W., Papageorgiou, A.C., Chronopoulou, E., Labrou, N.E.(2015) Biochim Biophys Acta 1854: 166-177
- PubMed: 25479053
- DOI: https://doi.org/10.1016/j.bbapap.2014.11.008
- Primary Citation of Related Structures:
4CHS - PubMed Abstract:
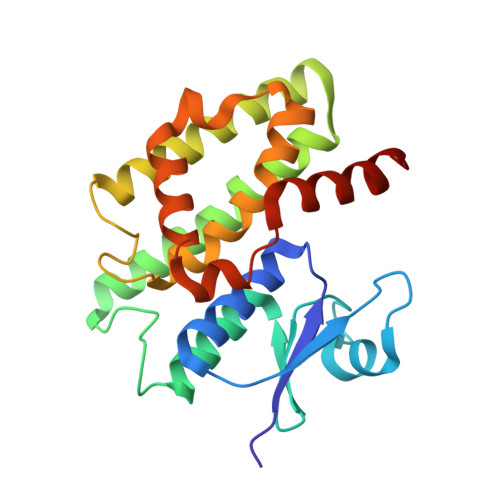
The plant tau class glutathione transferases (GSTs) play important roles in biotic and abiotic stress tolerance in crops and weeds. In this study, we systematically examined the catalytic and structural features of a GST isoenzyme from Glycine max (GmGSTU10-10). GmGSTU10-10 is a unique isoenzyme in soybean that is specifically expressed in response to biotic stress caused by soybean mosaic virus (SMV) infections. GmGSTU10-10 was cloned, expressed in Escherichia coli, purified and characterized. The results showed that GmGSTU10-10 catalyzes several different reactions and exhibits wide substrate specificity. Of particular importance is the finding that the enzyme shows high antioxidant catalytic function and acts as hydroperoxidase. In addition, its Km for GSH is significantly lower, compared to other plant GSTs, suggesting that GmGSTU10-10 is able to perform efficient catalysis under conditions where the concentration of reduced glutathione is low (e.g. oxidative stress). The crystal structure of GmGSTU10-10 was solved by molecular replacement at 1.6Å resolution in complex with glutathione sulfenic acid (GSOH). Structural analysis showed that GmGSTU10-10 shares the same overall fold and domain organization as other plant cytosolic GSTs; however, major variations were identified in helix H9 and the upper part of helix H4 that affect the size of the active site pockets, substrate recognition and the catalytic mechanism. The results of the present study provide new information into GST diversity and give further insights into the complex regulation and enzymatic functions of this plant gene superfamily.
- Laboratory of Enzyme Technology, Department of Biotechnology, School of Food, Biotechnology and Development, Agricultural University of Athens, 75 Iera Odos Street, GR-11855-Athens, Greece.
Organizational Affiliation: