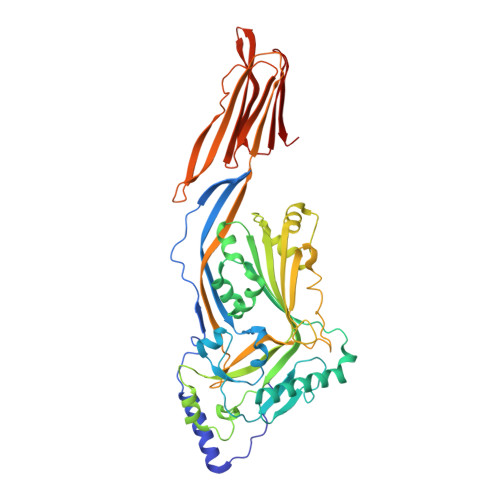
Crystal Structure of Listeriolysin O Reveals Molecular Details of Oligomerization and Pore Formation
Koester, S., Pee, K.V., Hudel, M., Leustik, M., Rhinow, D., Kuehlbrandt, W., Chakraborty, T., Yildiz, O.(2014) Nat Commun 5: 3690
- PubMed: 24751541
- DOI: https://doi.org/10.1038/ncomms4690
- Primary Citation of Related Structures:
4CDB - PubMed Abstract:
Listeriolysin O (LLO) is an essential virulence factor of Listeria monocytogenes that causes listeriosis. Listeria monocytogenes owes its ability to live within cells to the pH- and temperature-dependent pore-forming activity of LLO, which is unique among cholesterol-dependent cytolysins. LLO enables the bacteria to cross the phagosomal membrane and is also involved in activation of cellular processes, including the modulation of gene expression or intracellular Ca(2+) oscillations. Neither the pore-forming mechanism nor the mechanisms triggering the signalling processes in the host cell are known in detail. Here, we report the crystal structure of LLO, in which we identified regions important for oligomerization and pore formation. Mutants were characterized by determining their haemolytic and Ca(2+) uptake activity. We analysed the pore formation of LLO and its variants on erythrocyte ghosts by electron microscopy and show that pore formation requires precise interface interactions during toxin oligomerization on the membrane.
- Department of Structural Biology, Max Planck Institute of Biophysics, Max von Laue Str. 3, Frankfurt am Main 60438, Germany.
Organizational Affiliation: