Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Dobritzsch, D., Wang, H., Schneider, G., Yu, S.(2014) Biochem J 462: 441
- PubMed: 24947135
- DOI: https://doi.org/10.1042/BJ20140382
- Primary Citation of Related Structures:
4C5Y, 4C5Z, 4C60, 4C65 - PubMed Abstract:
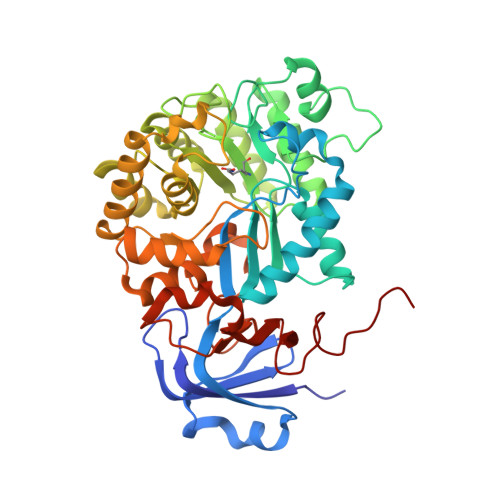
Ochratoxin, with ochratoxin A as the dominant form, is one of the five major mycotoxins most harmful to humans and animals. It is produced by Aspergillus and Penicillium species and occurs in a wide range of agricultural products. Detoxification of contaminated food is a challenging health issue. In the present paper we report the identification, characterization and crystal structure (at 2.2 Å) of a novel microbial ochratoxinase from Aspergillus niger. A putative amidase gene encoding a 480 amino acid polypeptide was cloned and homologously expressed in A. niger. The recombinant protein is N-terminally truncated, thermostable, has optimal activity at pH ~6 and 66°C, and is more efficient in ochratoxin A hydrolysis than carboxypeptidase A and Y, the two previously known enzymes capable of degrading this mycotoxin. The subunit of the homo-octameric enzyme folds into a two-domain structure characteristic of a metal dependent amidohydrolase, with a twisted TIM (triosephosphateisomerase)-barrel and a smaller β-sandwich domain. The active site contains an aspartate residue for acid-base catalysis, and a carboxylated lysine and four histidine residues for binding of a binuclear metal centre.
- *Department of Medical Biochemistry and Biophysics, Karolinska Institutet, SE-17177 Stockholm, Sweden.
Organizational Affiliation: