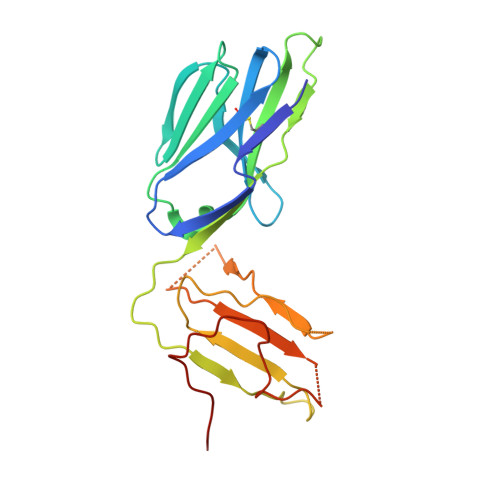
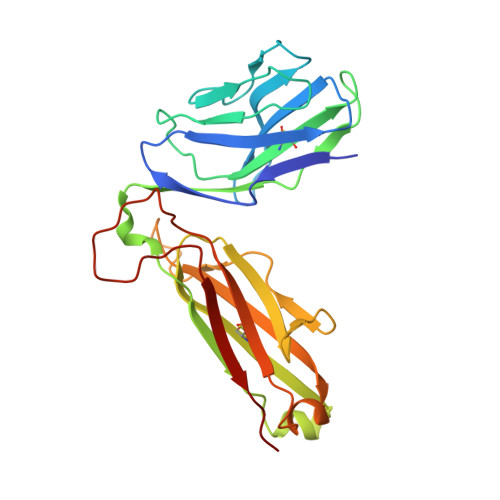
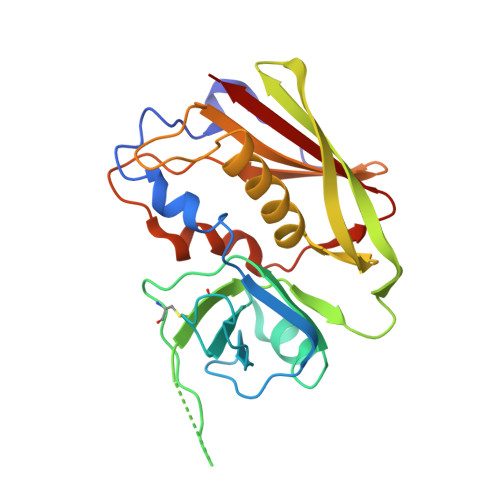
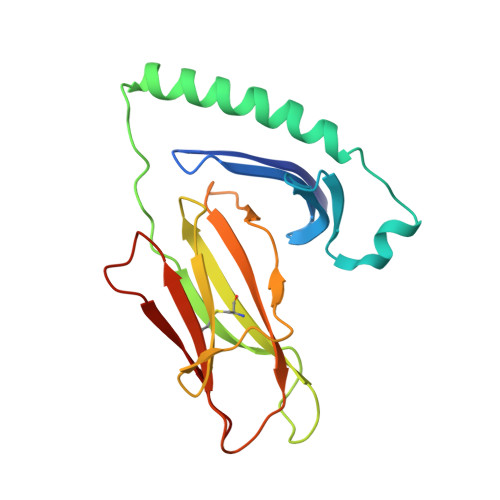
Structure of the Superantigen Staphylococcal Enterotoxin B in Complex with Tcr and Peptide-Mhc Demonstrates Absence of Tcr-Peptide Contacts.
Rodstrom, K.E.J., Elbing, K., Lindkvist-Petersson, K.(2014) J Immunol 193: 1998
- PubMed: 25015819
- DOI: https://doi.org/10.4049/jimmunol.1401268
- Primary Citation of Related Structures:
4C56 - PubMed Abstract:
Superantigens are immune-stimulatory toxins produced by Staphylococcus aureus, which are able to interact with host immune receptors to induce a massive release of cytokines, causing toxic shock syndrome and possibly death. In this article, we present the x-ray structure of staphylococcal enterotoxin B (SEB) in complex with its receptors, the TCR and MHC class II, forming a ternary complex. The structure, in combination with functional analyses, clearly shows how SEB adopts a wedge-like position when binding to the β-chain of TCR, allowing for an interaction between the α-chain of TCR and MHC. Furthermore, the binding mode also circumvents contact between TCR and the peptide presented by MHC, which enables SEB to initiate a peptide-independent activation of T cells.
- Department of Experimental Medical Science, Lund University, 221 84 Lund, Sweden.
Organizational Affiliation: