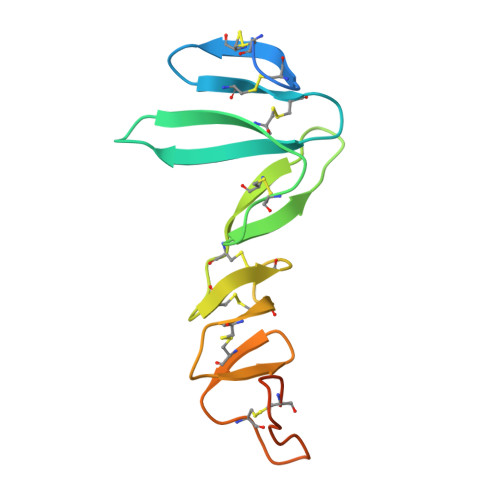
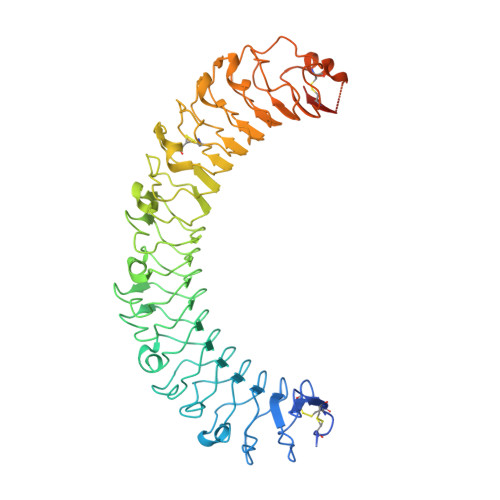Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Peng, W.C., De Lau, W., Forneris, F., Granneman, J.C.M., Huch, M., Clevers, H., Gros, P.(2013) Cell Rep 3: 1885
- PubMed: 23809763
- DOI: https://doi.org/10.1016/j.celrep.2013.06.009
- Primary Citation of Related Structures:
4BSO, 4BSP, 4BSR, 4BSS, 4BST, 4BSU - PubMed Abstract:
Leucine-rich repeat-containing G protein-coupled receptors 4-6 (LGR4-LGR6) are receptors for R-spondins, potent Wnt agonists that exert profound trophic effects on Wnt-driven stem cells compartments. We present crystal structures of a signaling-competent fragment of R-spondin 1 (Rspo1) at a resolution of 2.0 Å and its complex with the LGR5 ectodomain at a resolution of 3.2 Å. Ecto-LGR5 binds Rspo1 at its concave leucine-rich-repeat (LRR) surface, forming a dimeric 2:2 complex. Fully conserved residues on LGR4-LGR6 explain promiscuous binding of R-spondins. A phenylalanine clamp formed by Rspo1 Phe106 and Phe110 pinches Ala190 of LGR5 and is critical for binding. Mutations related to congenital anonychia reduce signaling, but not binding of Rspo1 to LGR5. Furthermore, antibody binding to the extended loop of the C-terminal LRR cap of LGR5 activates signaling in a ligand-independent manner. Thus, our data reveal binding of R-spondins to conserved sites on LGR4-LGR6 and, in analogy to FSHR and related receptors, suggest a direct signaling role for LGR4-LGR6 in addition to its formation of Wnt receptor and coreceptor complexes.
- Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Science, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands.
Organizational Affiliation: