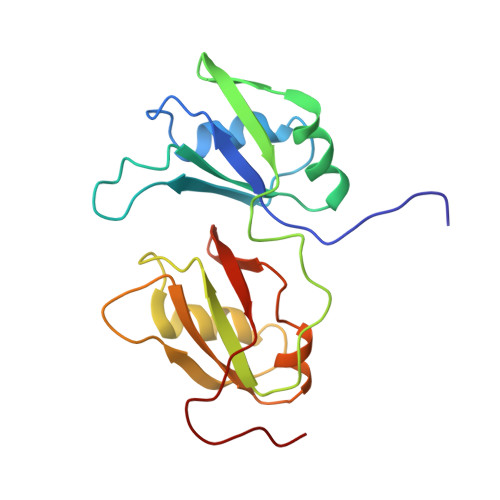
Molecular Basis of Ug-Rich RNA Recognition by the Human Splicing Factor Tdp-43
Lukavsky, P.J., Daujotyte, D., Tollervey, J.R., Ule, J., Stuani, C., Buratti, E., Baralle, F.E., Damberger, F.F., Allain, F.H.T.(2013) Nat Struct Mol Biol 20: 1443
- PubMed: 24240615
- DOI: https://doi.org/10.1038/nsmb.2698
- Primary Citation of Related Structures:
4BS2 - PubMed Abstract:
TDP-43 encodes an alternative-splicing regulator with tandem RNA-recognition motifs (RRMs). The protein regulates cystic fibrosis transmembrane regulator (CFTR) exon 9 splicing through binding to long UG-rich RNA sequences and is found in cytoplasmic inclusions of several neurodegenerative diseases. We solved the solution structure of the TDP-43 RRMs in complex with UG-rich RNA. Ten nucleotides are bound by both RRMs, and six are recognized sequence specifically. Among these, a central G interacts with both RRMs and stabilizes a new tandem RRM arrangement. Mutations that eliminate recognition of this key nucleotide or crucial inter-RRM interactions disrupt RNA binding and TDP-43-dependent splicing regulation. In contrast, point mutations that affect base-specific recognition in either RRM have weaker effects. Our findings reveal not only how TDP-43 recognizes UG repeats but also how RNA binding-dependent inter-RRM interactions are crucial for TDP-43 function.
- 1] Central European Institute of Technology, Masaryk University, Brno, Czech Republic. [2] Institute of Molecular Biology and Biophysics, Eidgenössische Technische Hochschule Hönggerberg, Zürich, Switzerland.
Organizational Affiliation: