Crystal Structure of the Aimp3-Mrs N-Terminal Domain Complex in Different Space Group
Cho, H.Y., Seo, W.W., Cho, H.J., Kang, B.S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| METHIONINE--TRNA LIGASE, CYTOPLASMIC | 232 | Homo sapiens | Mutation(s): 0 EC: 6.1.1.10 | 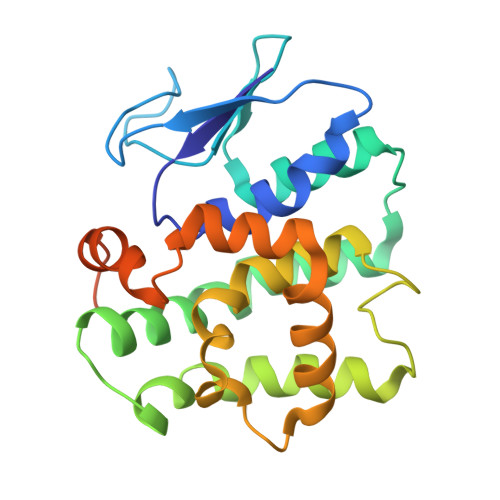 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P56192 (Homo sapiens) Explore P56192 Go to UniProtKB: P56192 | |||||
PHAROS: P56192 GTEx: ENSG00000166986 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P56192 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 EPSILON-1 | 186 | Homo sapiens | Mutation(s): 1 | 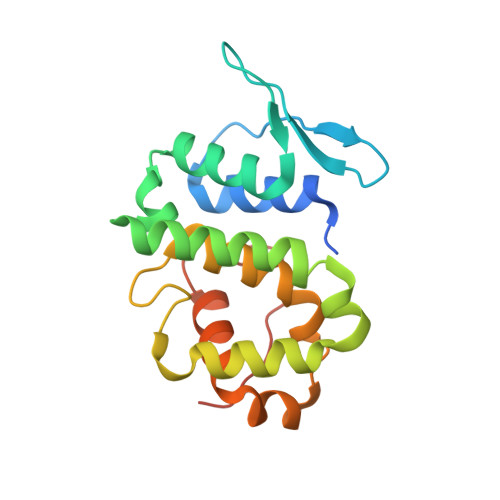 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O43324 (Homo sapiens) Explore O43324 Go to UniProtKB: O43324 | |||||
PHAROS: O43324 GTEx: ENSG00000124802 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O43324 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 43.685 | α = 90 |
| b = 72.029 | β = 90 |
| c = 123.417 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | phasing |