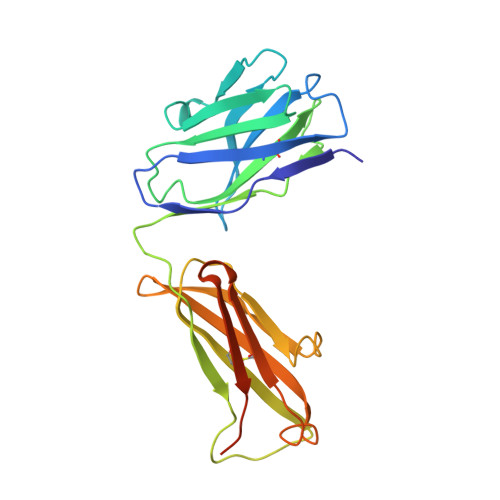
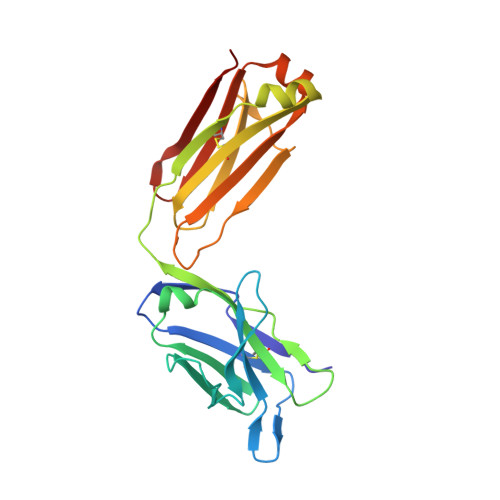
Epitope-Specific Antibody Response is Controlled by Immunoglobulin Vh Polymorphisms.
Raposo, B., Dobritzsch, D., Ge, C., Ekman, D., Xu, B., Lindh, I., Forster, M., Uysal, H., Nandakumar, K.S., Schneider, G., Holmdahl, R.(2014) J Exp Medicine 211: 405
- PubMed: 24534192
- DOI: https://doi.org/10.1084/jem.20130968
- Primary Citation of Related Structures:
4BKL - PubMed Abstract:
Autoantibody formation is essential for the development of certain autoimmune diseases like rheumatoid arthritis (RA). Anti-type II collagen (CII) antibodies are found in RA patients; they interact with cartilage in vivo and are often highly pathogenic in the mouse. Autoreactivity to CII is directed to multiple epitopes and conserved between mice and humans. We have previously mapped the antibody response to CII in a heterogeneous stock cohort of mice, with a strong association with the IgH locus. We positioned the genetic polymorphisms and determined the structural requirements controlling antibody recognition of one of the major CII epitopes. Polymorphisms at positions S31R and W33T of the associated variable heavy chain (VH) allele were identified and confirmed by gene sequencing. The Fab fragment binding the J1 epitope was crystallized, and site-directed mutagenesis confirmed the importance of those two variants for antigen recognition. Back mutation to germline sequence provided evidence for a preexisting recognition of the J1 epitope. These data demonstrate a genetic association of epitope-specific antibody responses with specific VH alleles, and it highlights the importance of germline-encoded antibodies in the pathogenesis of antibody-mediated autoimmune diseases.
- Section for Medical Inflammation Research and 2 Section for Molecular Structural Biology, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, 171 77 Stockholm, Sweden.
Organizational Affiliation: