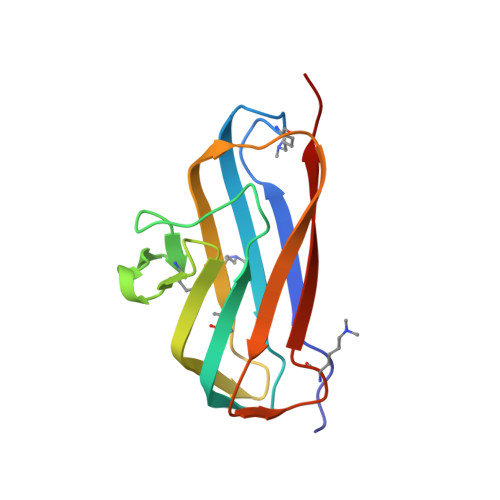
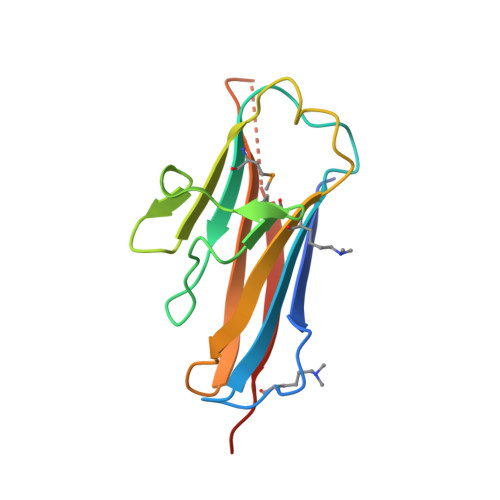
Bsla is a Self-Assembling Bacterial Hydrophobin that Coats the Bacillus Subtilis Biofilm.
Hobley, L., Ostrowski, A., Rao, F.V., Bromley, K.M., Porter, M., Prescott, A.R., Macphee, C.E., Van Aalten, D.M.F., Stanley-Wall, N.R.(2013) Proc Natl Acad Sci U S A 110: 13600
- PubMed: 23904481
- DOI: https://doi.org/10.1073/pnas.1306390110
- Primary Citation of Related Structures:
4BHU - PubMed Abstract:
Biofilms represent the predominant mode of microbial growth in the natural environment. Bacillus subtilis is a ubiquitous Gram-positive soil bacterium that functions as an effective plant growth-promoting agent. The biofilm matrix is composed of an exopolysaccharide and an amyloid fiber-forming protein, TasA, and assembles with the aid of a small secreted protein, BslA. Here we show that natively synthesized and secreted BslA forms surface layers around the biofilm. Biophysical analysis demonstrates that BslA can self-assemble at interfaces, forming an elastic film. Molecular function is revealed from analysis of the crystal structure of BslA, which consists of an Ig-type fold with the addition of an unusual, extremely hydrophobic "cap" region. A combination of in vivo biofilm formation and in vitro biophysical analysis demonstrates that the central hydrophobic residues of the cap are essential to allow a hydrophobic, nonwetting biofilm to form as they control the surface activity of the BslA protein. The hydrophobic cap exhibits physiochemical properties remarkably similar to the hydrophobic surface found in fungal hydrophobins; thus, BslA is a structurally defined bacterial hydrophobin. We suggest that biofilms formed by other species of bacteria may have evolved similar mechanisms to provide protection to the resident bacterial community.
- Division of Molecular Microbiology, College of Life Sciences, University of Dundee, Dundee DD1 5EH, United Kingdom.
Organizational Affiliation: