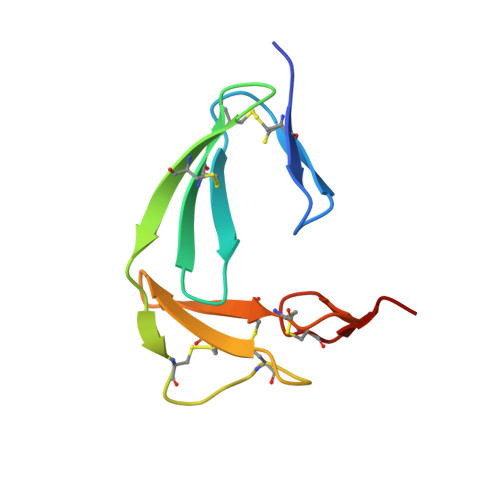
The Structure of the Fni-Egf-Like Tandem Domain of Coagulation Factor Xii Solved Using Siras.
Beringer, D.X., Kroon-Batenburg, L.M.J.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 94
- PubMed: 23385745
- DOI: https://doi.org/10.1107/S1744309113000286
- Primary Citation of Related Structures:
4BDW, 4BDX - PubMed Abstract:
Coagulation factor XII (FXII) is a key protein in the intrinsic coagulation and kallikrein-kinin pathways. It has been found that negative surfaces and amyloids, such as Aβ fibrils, can activate FXII. Additionally, it has been suggested that FXII simulates cells and that it plays an important role in thrombosis. To date, no structural data on FXII have been deposited, which makes it difficult to support any hypothesis on the mechanism of FXII function. The crystal structure of the FnI-EGF-like tandem domain of FXII presented here was solved using experimental phases. To determine the phases, a SIRAS approach was used with a native and a holmium chloride-soaked data set. The holmium cluster was coordinated by the C-terminal tails of two symmetry-related molecules. Another observation was that the FnI domain was much more ordered than the EGF-like domain owing to crystal packing. Furthermore, the structure shows the same domain orientation as the homologous FnI-EGF-like tandem domain of tPA. The plausibility of several proposed interactions of these domains of FXII is discussed. Based on this FXII FnI-EGF-like structure, it could be possible that FXII binding to amyloid and negatively charged surfaces is mediated via this part of FXII.
- Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Department of Chemistry, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands.
Organizational Affiliation: