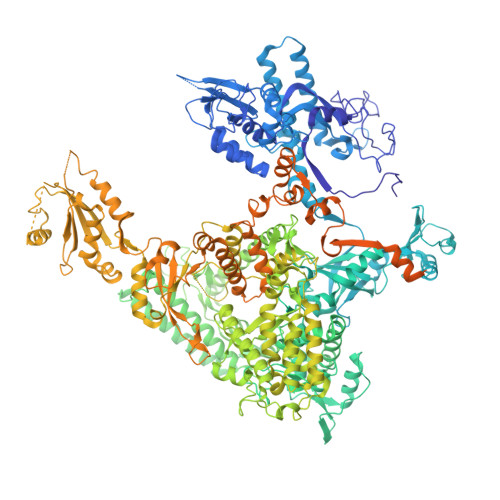
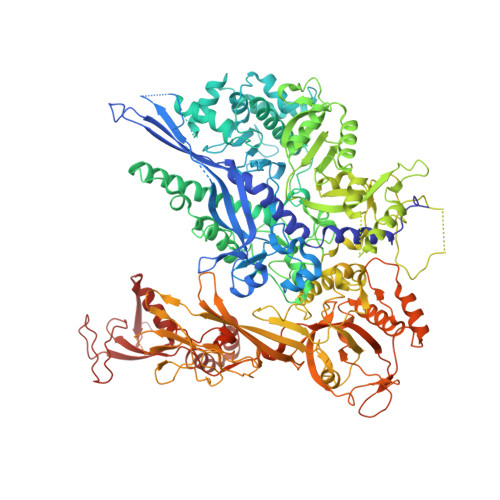
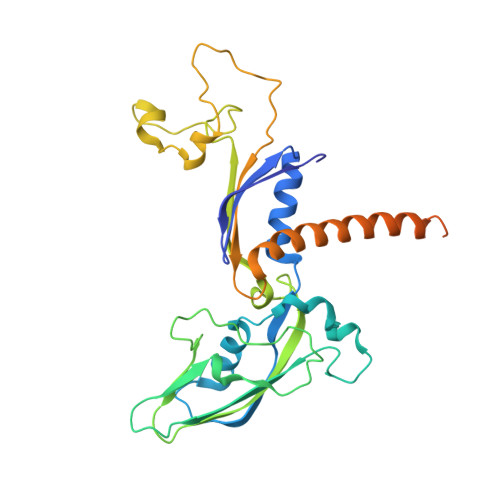
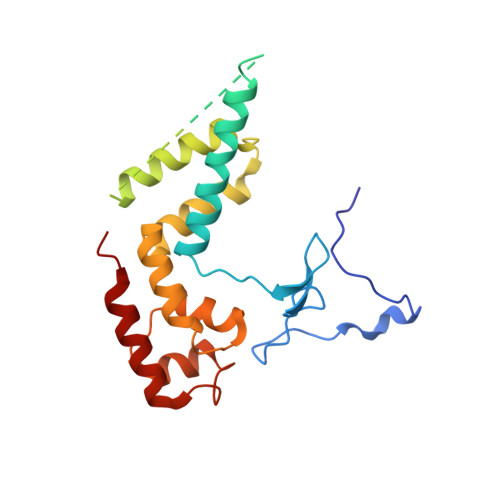
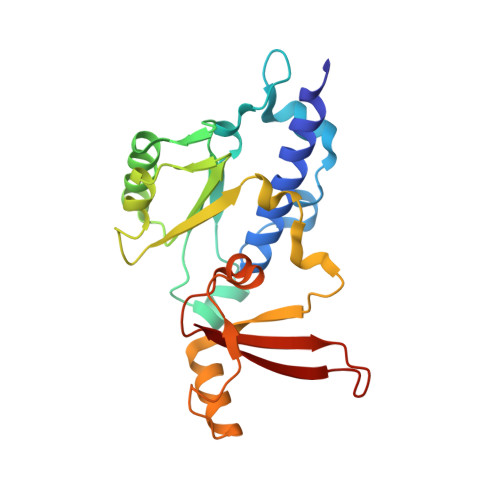
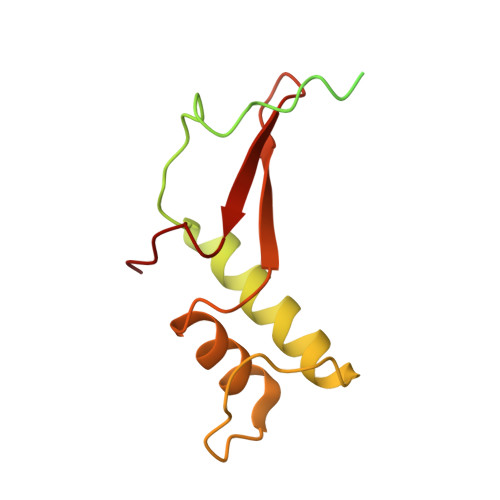
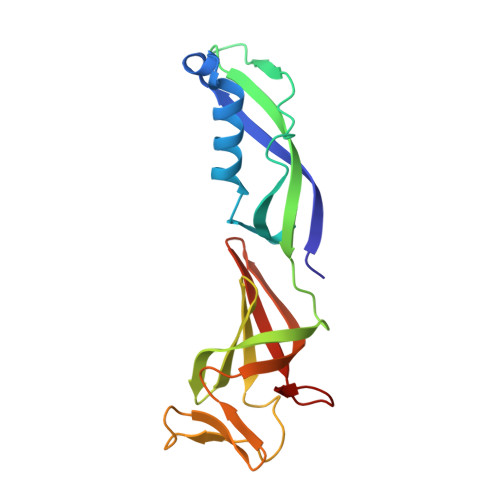
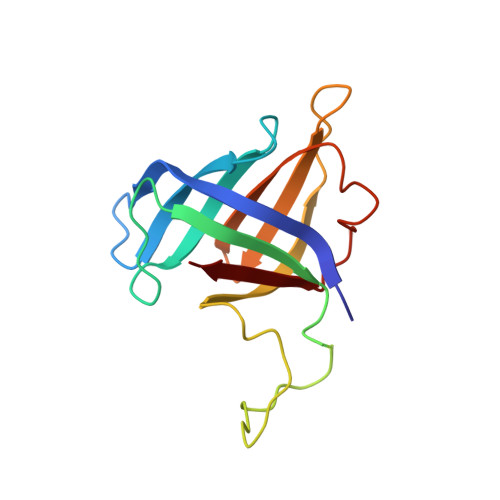
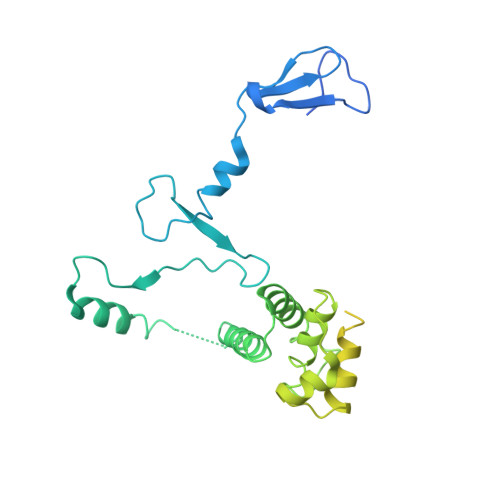
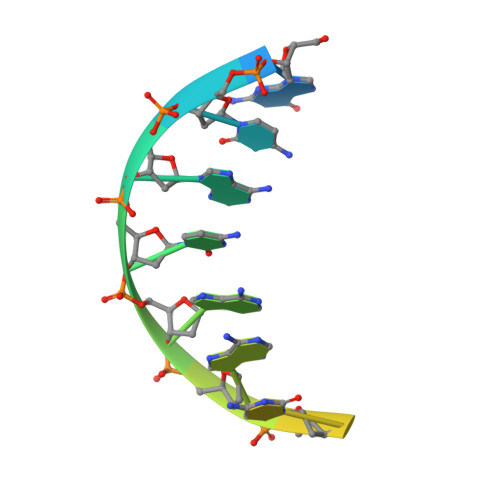
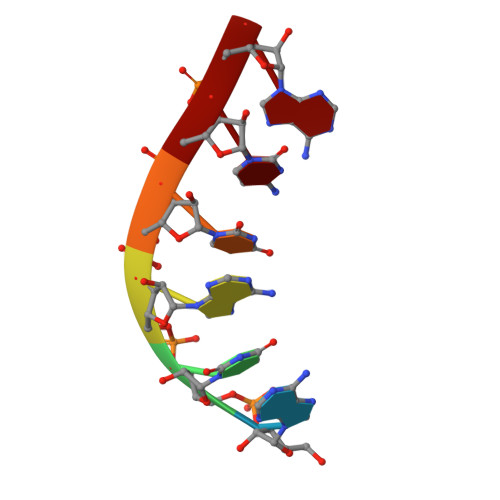
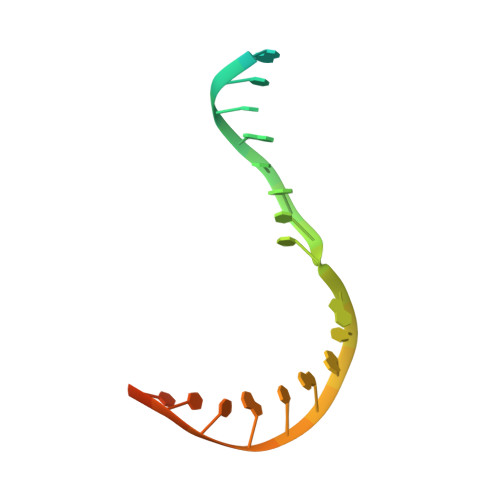Structure and Function of the Initially Transcribing RNA Polymerase II-TFIIB Complex
Sainsbury, S., Niesser, J., Cramer, P.(2013) Nature 493: 437
- PubMed: 23151482
- DOI: https://doi.org/10.1038/nature11715
- Primary Citation of Related Structures:
4BBR, 4BBS - PubMed Abstract:
The general transcription factor (TF) IIB is required for RNA polymerase (Pol) II initiation and extends with its B-reader element into the Pol II active centre cleft. Low-resolution structures of the Pol II-TFIIB complex indicated how TFIIB functions in DNA recruitment, but they lacked nucleic acids and half of the B-reader, leaving other TFIIB functions enigmatic. Here we report crystal structures of the Pol II-TFIIB complex from the yeast Saccharomyces cerevisiae at 3.4 Å resolution and of an initially transcribing complex that additionally contains the DNA template and a 6-nucleotide RNA product. The structures reveal the entire B-reader and protein-nucleic acid interactions, and together with functional data lead to a more complete understanding of transcription initiation. TFIIB partially closes the polymerase cleft to position DNA and assist in its opening. The B-reader does not reach the active site but binds the DNA template strand upstream to assist in the recognition of the initiator sequence and in positioning the transcription start site. TFIIB rearranges active-site residues, induces binding of the catalytic metal ion B, and stimulates initial RNA synthesis allosterically. TFIIB then prevents the emerging DNA-RNA hybrid duplex from tilting, which would impair RNA synthesis. When the RNA grows beyond 6 nucleotides, it is separated from DNA and is directed to its exit tunnel by the B-reader loop. Once the RNA grows to 12-13 nucleotides, it clashes with TFIIB, triggering TFIIB displacement and elongation complex formation. Similar mechanisms may underlie all cellular transcription because all eukaryotic and archaeal RNA polymerases use TFIIB-like factors, and the bacterial initiation factor sigma has TFIIB-like topology and contains the loop region 3.2 that resembles the B-reader loop in location, charge and function. TFIIB and its counterparts may thus account for the two fundamental properties that distinguish RNA from DNA polymerases: primer-independent chain initiation and product separation from the template.
- Gene Center and Department of Biochemistry, Center for Integrated Protein Science CIPSM, Ludwig-Maximilians-Universität München, Feodor-Lynen-Str. 25, 81377 Munich, Germany.
Organizational Affiliation: