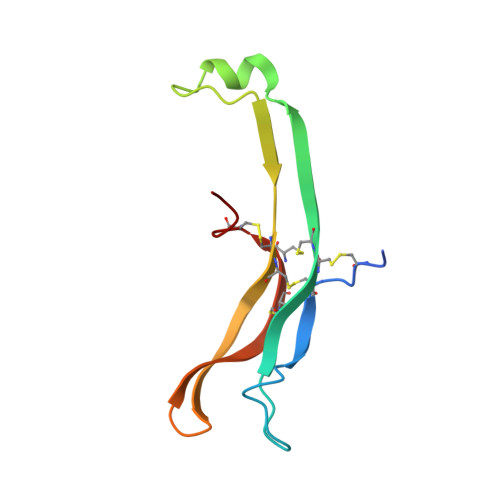
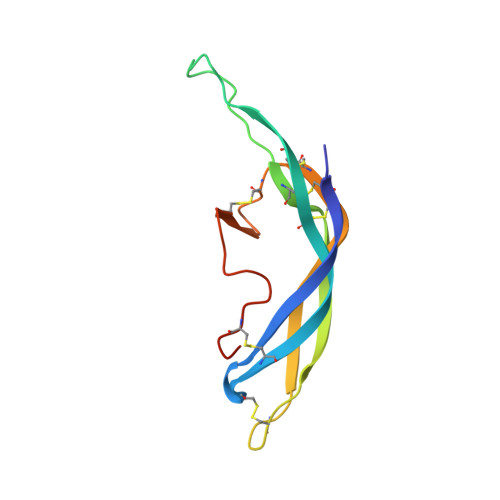
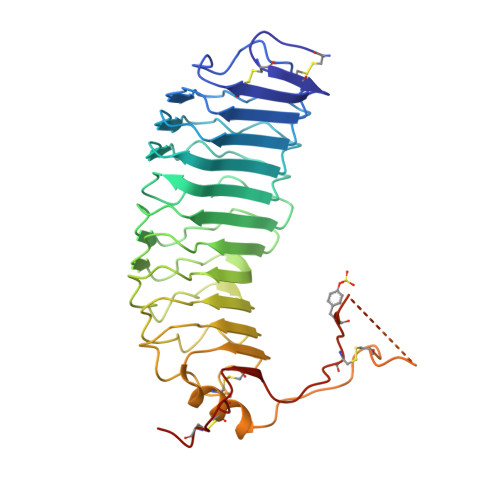
Structure of Follicle-Stimulating Hormone in Complex with the Entire Ectodomain of its Receptor.
Jiang, X., Liu, H., Chen, X., Chen, P., Fischer, D., Sriraman, V., Yu, H.N., Arkinstall, S., He, X.(2012) Proc Natl Acad Sci U S A 109: 12491
- PubMed: 22802634
- DOI: https://doi.org/10.1073/pnas.1206643109
- Primary Citation of Related Structures:
4AY9 - PubMed Abstract:
FSH, a glycoprotein hormone, and the FSH receptor (FSHR), a G protein-coupled receptor, play central roles in human reproduction. We report the crystal structure of FSH in complex with the entire extracellular domain of FSHR (FSHR(ED)), including the enigmatic hinge region that is responsible for signal specificity. Surprisingly, the hinge region does not form a separate structural unit as widely anticipated but is part of the integral structure of FSHR(ED). In addition to the known hormone-binding site, FSHR(ED) provides interaction sites with the hormone: a sulfotyrosine (sTyr) site in the hinge region consistent with previous studies and a potential exosite resulting from putative receptor trimerization. Our structure, in comparison to others, suggests FSHR interacts with its ligand in two steps: ligand recruitment followed by sTyr recognition. FSH first binds to the high-affinity hormone-binding subdomain of FSHR and reshapes the ligand conformation to form a sTyr-binding pocket. FSHR then inserts its sTyr (i.e., sulfated Tyr335) into the FSH nascent pocket, eventually leading to receptor activation.
- EMD Serono Research Institute, Billerica, MA 01821, USA.
Organizational Affiliation: