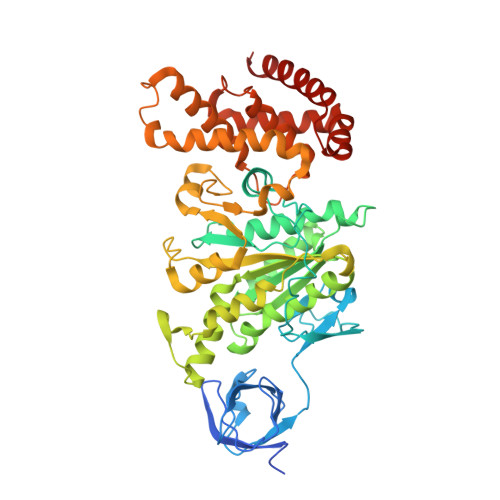
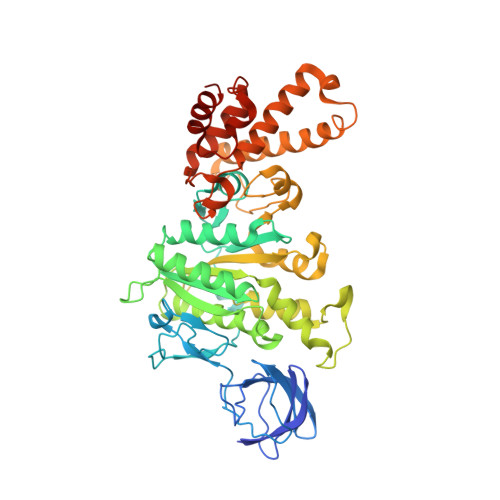
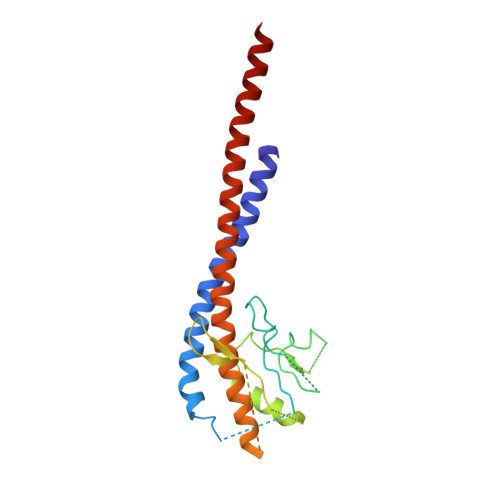
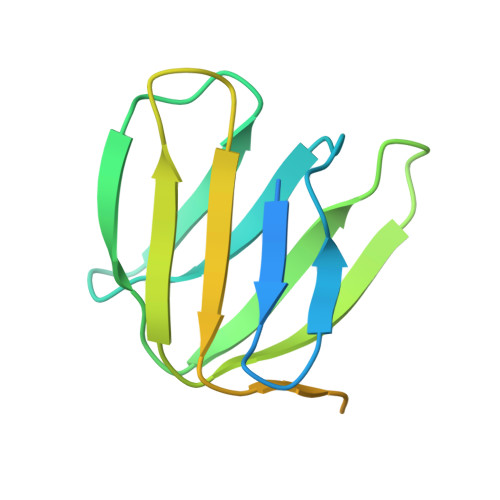
Structural Evidence of a New Catalytic Intermediate in the Pathway of ATP Hydrolysis by F1-ATPase from Bovine Heart Mitochondria.
Rees, D.M., Montgomery, M.G., Leslie, A.G., Walker, J.E.(2012) Proc Natl Acad Sci U S A 109: 11139
- PubMed: 22733764
- DOI: https://doi.org/10.1073/pnas.1207587109
- Primary Citation of Related Structures:
4ASU - PubMed Abstract:
The molecular description of the mechanism of F(1)-ATPase is based mainly on high-resolution structures of the enzyme from mitochondria, coupled with direct observations of rotation in bacterial enzymes. During hydrolysis of ATP, the rotor turns counterclockwise (as viewed from the membrane domain of the intact enzyme) in 120° steps. Because the rotor is asymmetric, at any moment the three catalytic sites are at different points in the catalytic cycle. In a "ground-state" structure of the bovine enzyme, one site (β(E)) is devoid of nucleotide and represents a state that has released the products of ATP hydrolysis. A second site (β(TP)) has bound the substrate, magnesium. ATP, in a precatalytic state, and in the third site (β(DP)), the substrate is about to undergo hydrolysis. Three successive 120° turns of the rotor interconvert the sites through these three states, hydrolyzing three ATP molecules, releasing the products and leaving the enzyme with two bound nucleotides. A transition-state analog structure, F(1)-TS, displays intermediate states between those observed in the ground state. For example, in the β(DP)-site of F(1)-TS, the terminal phosphate of an ATP molecule is undergoing in-line nucleophilic attack by a water molecule. As described here, we have captured another intermediate in the catalytic cycle, which helps to define the order of substrate release. In this structure, the β(E)-site is occupied by the product ADP, but without a magnesium ion or phosphate, providing evidence that the nucleotide is the last of the products of ATP hydrolysis to be released.
- Medical Research Council Mitochondrial Biology Unit, Cambridge CB2 2XY, United Kingdom.
Organizational Affiliation: