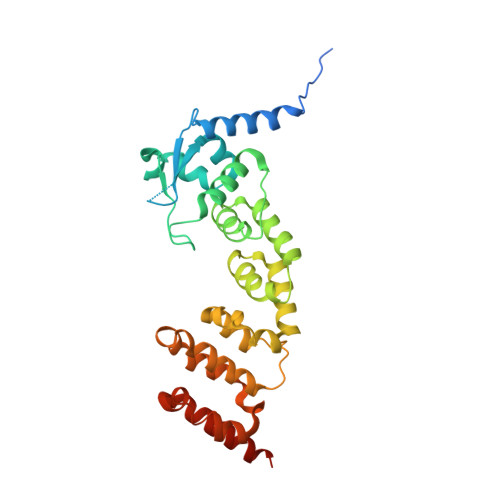
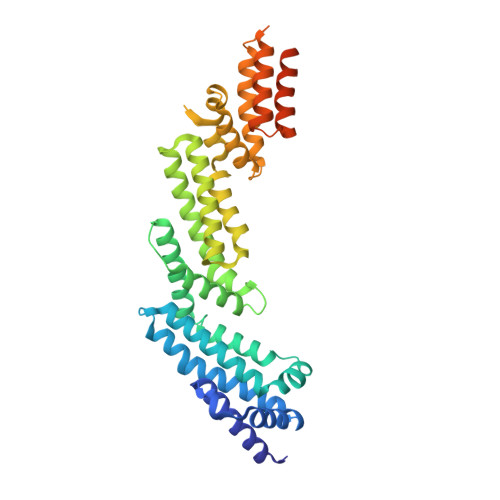
Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
Canning, P., Cooper, C.D.O., Krojer, T., Murray, J.W., Pike, A.C.W., Chaikuad, A., Keates, T., Thangaratnarajah, C., Hojzan, V., Marsden, B.D., Gileadi, O., Knapp, S., von Delft, F., Bullock, A.N.(2013) J Biological Chem 288: 7803
- PubMed: 23349464
- DOI: https://doi.org/10.1074/jbc.M112.437996
- Primary Citation of Related Structures:
2VPJ, 2XN4, 3II7, 4AP2, 4APF, 4ASC - PubMed Abstract:
Cullin-RING ligases are multisubunit E3 ubiquitin ligases that recruit substrate-specific adaptors to catalyze protein ubiquitylation. Cul3-based Cullin-RING ligases are uniquely associated with BTB adaptors that incorporate homodimerization, Cul3 assembly, and substrate recognition into a single multidomain protein, of which the best known are BTB-BACK-Kelch domain proteins, including KEAP1. Cul3 assembly requires a BTB protein "3-box" motif, analogous to the F-box and SOCS box motifs of other Cullin-based E3s. To define the molecular basis for this assembly and the overall architecture of the E3, we determined the crystal structures of the BTB-BACK domains of KLHL11 both alone and in complex with Cul3, along with the Kelch domain structures of KLHL2 (Mayven), KLHL7, KLHL12, and KBTBD5. We show that Cul3 interaction is dependent on a unique N-terminal extension sequence that packs against the 3-box in a hydrophobic groove centrally located between the BTB and BACK domains. Deletion of this N-terminal region results in a 30-fold loss in affinity. The presented data offer a model for the quaternary assembly of this E3 class that supports the bivalent capture of Nrf2 and reveals potential new sites for E3 inhibitor design.
- Structural Genomics Consortium, University of Oxford, Oxford OX3 7DQ, United Kingdom.
Organizational Affiliation: