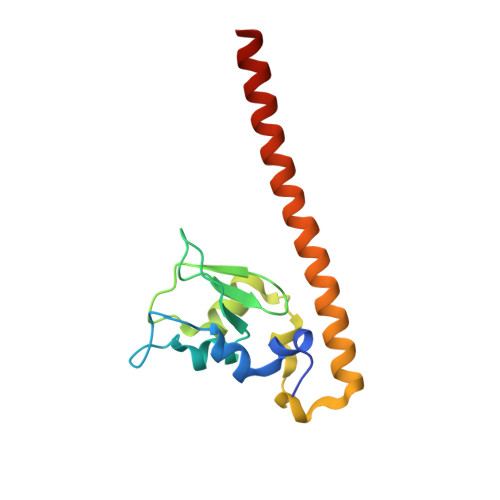
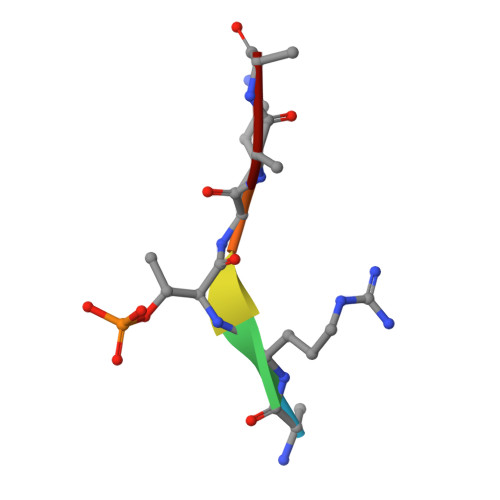
Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Jeyaprakash, A.A., Basquin, C., Jayachandran, U., Conti, E.(2011) Structure 19: 1625
- PubMed: 22032967
- DOI: https://doi.org/10.1016/j.str.2011.09.002
- Primary Citation of Related Structures:
4A0I, 4A0J, 4A0N - PubMed Abstract:
Localization of the chromosomal passenger complex (CPC) at centromeres during early mitosis is essential for accurate chromosome segregation and is dependent on the phosphorylation of histone H3. We report the 2.7 Å resolution structure of the CPC subunit Survivin bound to the N-terminal tail of histone H3 carrying the Thr3 phosphorylation mark (Thr3ph). The BIR domain of Survivin recognizes the Ala1-Arg2-Thr3ph-Lys4 sequence, decoding the modification state and the free N terminus of histone H3 by a strategy similar to that used by PHD fingers. The structural analysis permitted the identification of putative Survivin-binding epitopes in other mitotic proteins, including human Shugoshin 1. Using biophysical and structural data, we show that a phospho-mimic N-terminal sequence such as that of hSgo1 (Ala1-Lys2-Glu3-Arg4) contains the specificity determinants to bind Survivin. Our findings suggest that the CPC engages in mutually exclusive interactions with other constituents of the mitotic machinery and a histone mark in chromatin.
- Department of Structural Cell Biology, Max-Planck-Institute of Biochemistry, Am Klopferspitz 18, D-82152 Martinsried, Germany. jeyaprak@biochem.mpg.de
Organizational Affiliation: