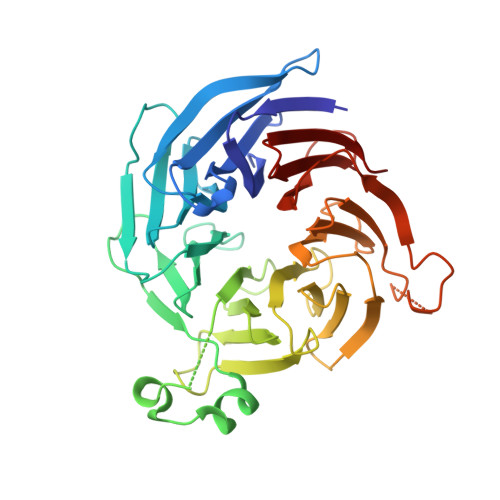
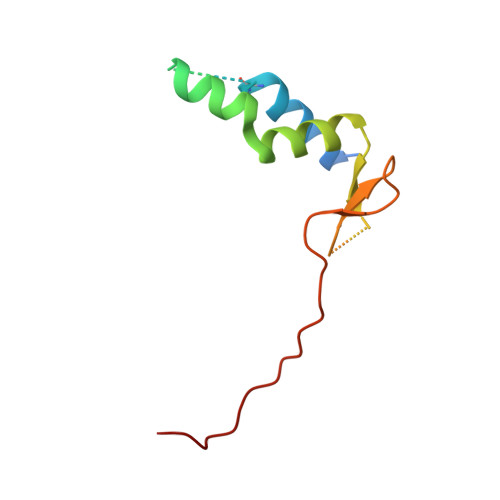
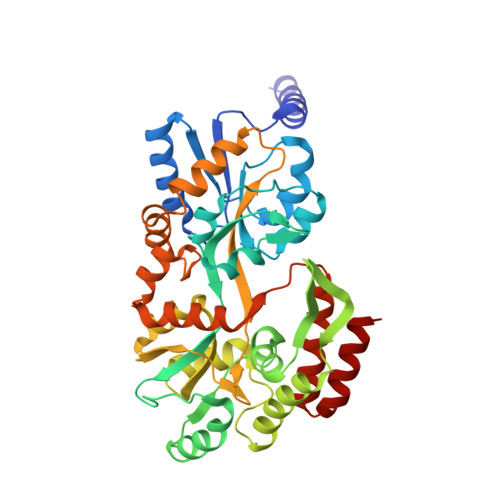
Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p
Pan, D., Nakatsu, T., Kato, H.(2013) Nat Struct Mol Biol 20: 987-993
- PubMed: 23812376
- DOI: https://doi.org/10.1038/nsmb.2618
- Primary Citation of Related Structures:
3W15 - PubMed Abstract:
Appropriate targeting of matrix proteins to peroxisomes is mainly directed by two types of peroxisomal targeting signals, PTS1 and PTS2. Although the basis of PTS1 recognition has been revealed by structural studies, that of PTS2 recognition remains elusive. Here we present the crystal structure of a heterotrimeric PTS2-recognition complex from Saccharomyces cerevisiae, containing Pex7p, the C-terminal region of Pex21p and the PTS2 of the peroxisomal 3-ketoacyl-CoA thiolase. Pex7p forms a β-propeller structure and provides a platform for cooperative interactions with both the amphipathic PTS2 helix and Pex21p. The C-terminal region of Pex21p directly covers the hydrophobic surfaces of both Pex7p and PTS2, and the resulting hydrophobic core is the primary determinant of stable complex formation. Together with in vivo and in vitro functional assays of Pex7p and Pex21p variants, our findings reveal the molecular mechanism of PTS2 recognition.
- Department of Structural Biology, Graduate School of Pharmaceutical Sciences, Kyoto University, Kyoto, Japan. pan.dongqing@pharm.kyoto-u.ac.jp
Organizational Affiliation: