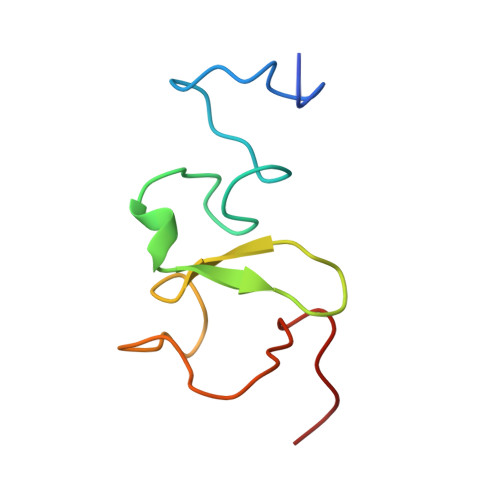
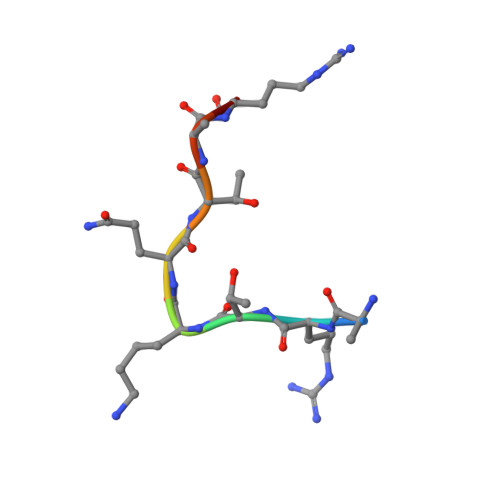
PHD Finger Recognition of Unmodified Histone H3R2 Links UHRF1 to Regulation of Euchromatic Gene Expression.
Rajakumara, E., Wang, Z., Ma, H., Hu, L., Chen, H., Lin, Y., Guo, R., Wu, F., Li, H., Lan, F., Shi, Y.G., Xu, Y., Patel, D.J., Shi, Y.(2011) Mol Cell 43: 275-284
- PubMed: 21777816
- DOI: https://doi.org/10.1016/j.molcel.2011.07.006
- Primary Citation of Related Structures:
3SOU, 3SOW, 3SOX - PubMed Abstract:
Histone methylation occurs on both lysine and arginine residues, and its dynamic regulation plays a critical role in chromatin biology. Here we identify the UHRF1 PHD finger (PHD(UHRF1)), an important regulator of DNA CpG methylation, as a histone H3 unmodified arginine 2 (H3R2) recognition modality. This conclusion is based on binding studies and cocrystal structures of PHD(UHRF1) bound to histone H3 peptides, where the guanidinium group of unmodified R2 forms an extensive intermolecular hydrogen bond network, with methylation of H3R2, but not H3K4 or H3K9, disrupting complex formation. We have identified direct target genes of UHRF1 from microarray and ChIP studies. Importantly, we show that UHRF1's ability to repress its direct target gene expression is dependent on PHD(UHRF1) binding to unmodified H3R2, thereby demonstrating the functional importance of this recognition event and supporting the potential for crosstalk between histone arginine methylation and UHRF1 function.
- Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10065, USA.
Organizational Affiliation: