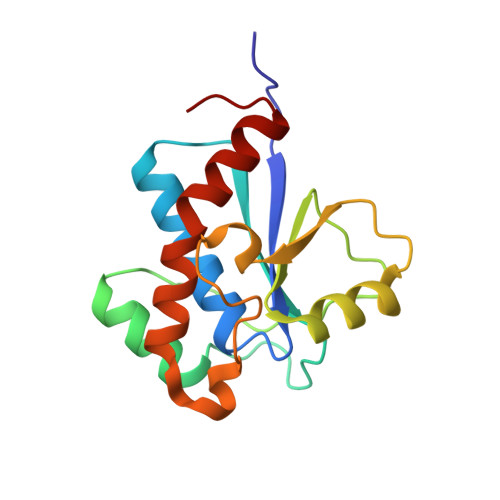
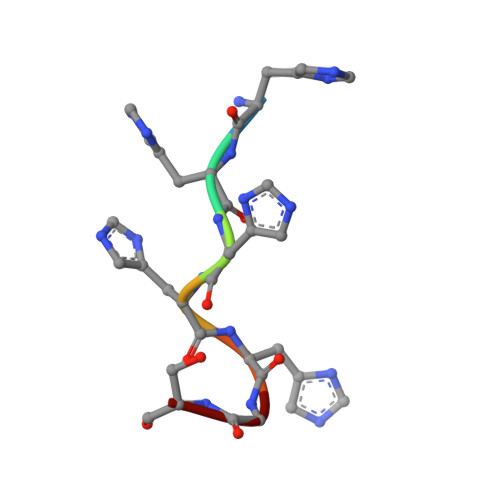
Structure and substrate recognition of the Staphylococcus aureus protein tyrosine phosphatase PtpA.
Vega, C., Chou, S., Engel, K., Harrell, M.E., Rajagopal, L., Grundner, C.(2011) J Mol Biology 413: 24-31
- PubMed: 21871460
- DOI: https://doi.org/10.1016/j.jmb.2011.08.015
- Primary Citation of Related Structures:
3ROF - PubMed Abstract:
Phosphosignaling through pSer/pThr/pTyr is emerging as a common signaling mechanism in prokaryotes. The human pathogen Staphylococcus aureus produces two low-molecular-weight protein tyrosine phosphatases (PTPs), PtpA and PtpB, with unknown functions. To provide the structural context for understanding PtpA function and substrate recognition, establish PtpA's structural relations within the PTP family, and provide a framework for the design of specific inhibitors, we solved the crystal structure of PtpA at 1 Å resolution. While PtpA adopts the common, conserved PTP fold and shows close overall similarity to eukaryotic PTPs, several features in the active site and surface organization are unique and can be explored to design selective inhibitors. A peptide bound in the active site mimics a phosphotyrosine substrate, affords insight into substrate recognition, and provides a testable substrate prediction. Genetic deletion of ptpA or ptpB does not affect in vitro growth or cell wall integrity, raising the possibility that PtpA and PtpB have specialized functions during infection.
- Seattle Biomedical Research Institute, Seattle, WA 98109, USA.
Organizational Affiliation: