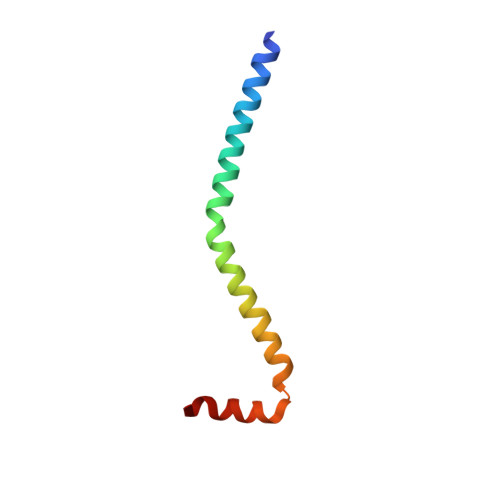
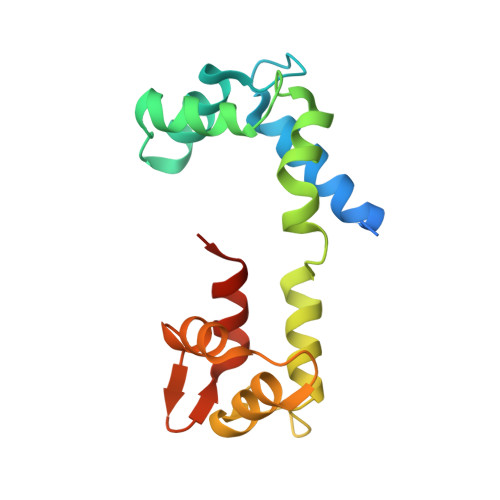
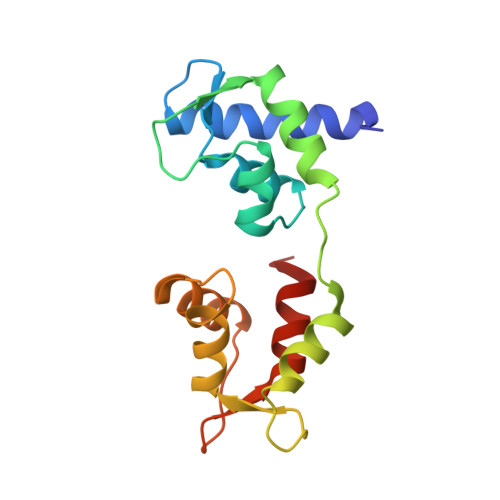
Visualizing key hinges and a potential major source of compliance in the lever arm of myosin.
Brown, J.H., Senthil Kumar, V.S., O'Neall-Hennessey, E., Reshetnikova, L., Robinson, H., Nguyen-McCarty, M., Szent-Gyorgyi, A.G., Cohen, C.(2011) Proc Natl Acad Sci U S A 108: 114-119
- PubMed: 21149681
- DOI: https://doi.org/10.1073/pnas.1016288107
- Primary Citation of Related Structures:
3PN7 - PubMed Abstract:
We have determined the 2.3-Å-resolution crystal structure of a myosin light chain domain, corresponding to one type found in sea scallop catch ("smooth") muscle. This structure reveals hinges that may function in the "on" and "off" states of myosin. The molecule adopts two different conformations about the heavy chain "hook" and regulatory light chain (RLC) helix D. This conformational change results in extended and compressed forms of the lever arm whose lengths differ by 10 Å. The heavy chain hook and RLC helix D hinges could thus serve as a potential major and localized source of cross-bridge compliance during the contractile cycle. In addition, in one of the molecules of the crystal, part of the RLC N-terminal extension is seen in atomic detail and forms a one-turn alpha-helix that interacts with RLC helix D. This extension, whose sequence is highly variable in different myosins, may thus modulate the flexibility of the lever arm. Moreover, the relative proximity of the phosphorylation site to the helix D hinge suggests a potential role for conformational changes about this hinge in the transition between the on and off states of regulated myosins.
- Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, MA 02454-9110, USA.
Organizational Affiliation: