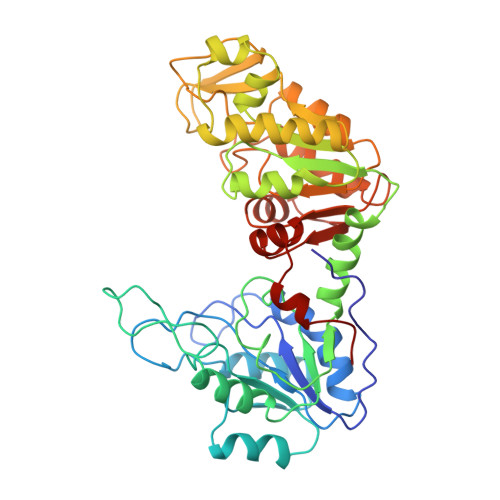
Sequence and structure of yeast phosphoglycerate kinase.
Watson, H.C., Walker, N.P., Shaw, P.J., Bryant, T.N., Wendell, P.L., Fothergill, L.A., Perkins, R.E., Conroy, S.C., Dobson, M.J., Tuite, M.F.(1982) EMBO J 1: 1635-1640
- PubMed: 6765200
- DOI: https://doi.org/10.1002/j.1460-2075.1982.tb01366.x
- Primary Citation of Related Structures:
3PGK - PubMed Abstract:
The structure of yeast phosphoglycerate kinase has been determined with data obtained from amino acid sequence, nucleotide sequence, and X-ray crystallographic studies. The substrate binding sites, as deduced from electron density maps, are compatible with known substrate specificity and the stereochemical requirements for the enzymic reaction. A carboxyl-imidazole interaction appears to be involved in controlling the transition between the open and closed forms of the enzyme.