Structural and Inhibition Studies of the RNase H Function of Xenotropic Murine Leukemia Virus-Related Virus Reverse Transcriptase.
Kirby, K.A., Marchand, B., Ong, Y.T., Ndongwe, T.P., Hachiya, A., Michailidis, E., Leslie, M.D., Sietsema, D.V., Fetterly, T.L., Dorst, C.A., Singh, K., Wang, Z., Parniak, M.A., Sarafianos, S.G.(2012) Antimicrob Agents Chemother 56: 2048-2061
- PubMed: 22252812
- DOI: https://doi.org/10.1128/AAC.06000-11
- Primary Citation of Related Structures:
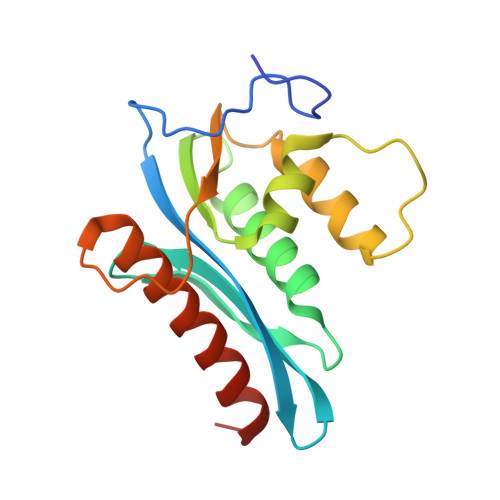
3P1G - PubMed Abstract:
RNase H inhibitors (RNHIs) have gained attention as potential HIV-1 therapeutics. Although several RNHIs have been studied in the context of HIV-1 reverse transcriptase (RT) RNase H, there is no information on inhibitors that might affect the RNase H activity of other RTs. We performed biochemical, virological, crystallographic, and molecular modeling studies to compare the RNase H function and inhibition profiles of the gammaretroviral xenotropic murine leukemia virus-related virus (XMRV) and Moloney murine leukemia virus (MoMLV) RTs to those of HIV-1 RT. The RNase H activity of XMRV RT is significantly lower than that of HIV-1 RT and comparable to that of MoMLV RT. XMRV and MoMLV, but not HIV-1 RT, had optimal RNase H activities in the presence of Mn²⁺ and not Mg²⁺. Using hydroxyl-radical footprinting assays, we demonstrated that the distance between the polymerase and RNase H domains in the MoMLV and XMRV RTs is longer than that in the HIV-1 RT by ∼3.4 Å. We identified one naphthyridinone and one hydroxyisoquinolinedione as potent inhibitors of HIV-1 and XMRV RT RNases H with 50% inhibitory concentrations ranging from ∼0.8 to 0.02 μM. Two acylhydrazones effective against HIV-1 RT RNase H were less potent against the XMRV enzyme. We also solved the crystal structure of an XMRV RNase H fragment at high resolution (1.5 Å) and determined the molecular details of the XMRV RNase H active site, thus providing a framework that would be useful for the design of antivirals that target RNase H.
- Christopher S. Bond Life Sciences Center, University of Missouri, Columbia, Missouri, USA.
Organizational Affiliation: