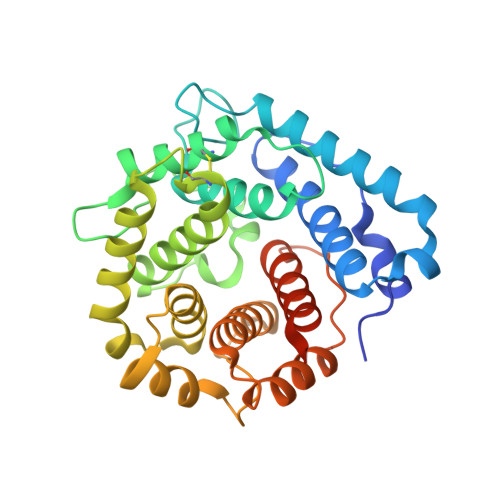
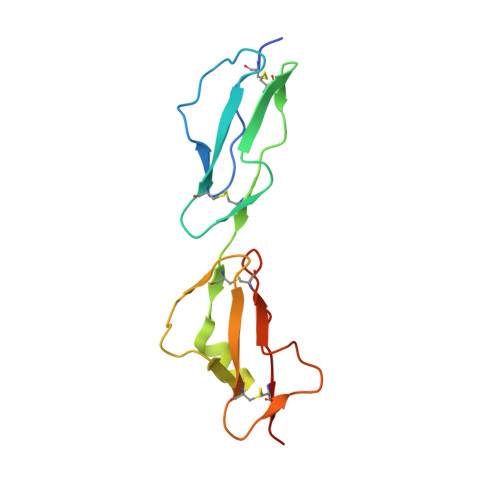
Structural basis for engagement by complement factor H of C3b on a self surface.
Morgan, H.P., Schmidt, C.Q., Guariento, M., Blaum, B.S., Gillespie, D., Herbert, A.P., Kavanagh, D., Mertens, H.D., Svergun, D.I., Johansson, C.M., Uhrin, D., Barlow, P.N., Hannan, J.P.(2011) Nat Struct Mol Biol 18: 463-470
- PubMed: 21317894
- DOI: https://doi.org/10.1038/nsmb.2018
- Primary Citation of Related Structures:
3OXU - PubMed Abstract:
Complement factor H (FH) attenuates C3b molecules tethered by their thioester domains to self surfaces and thereby protects host tissues. Factor H is a cofactor for initial C3b proteolysis that ultimately yields a surface-attached fragment (C3d) corresponding to the thioester domain. We used NMR and X-ray crystallography to study the C3d-FH19-20 complex in atomic detail and identify glycosaminoglycan-binding residues in factor H module 20 of the C3d-FH19-20 complex. Mutagenesis justified the merging of the C3d-FH19-20 structure with an existing C3b-FH1-4 crystal structure. We concatenated the merged structure with the available FH6-8 crystal structure and new SAXS-derived FH1-4, FH8-15 and FH15-19 envelopes. The combined data are consistent with a bent-back factor H molecule that binds through its termini to two sites on one C3b molecule and simultaneously to adjacent polyanionic host-surface markers.
- Institute of Structural and Molecular Biology, School of Biological Sciences, King's Buildings, University of Edinburgh, Edinburgh, UK.
Organizational Affiliation: