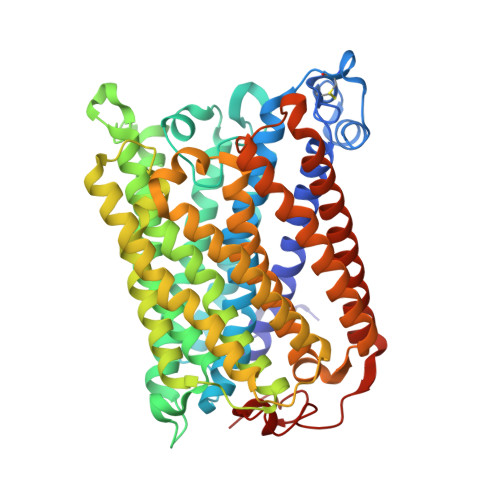
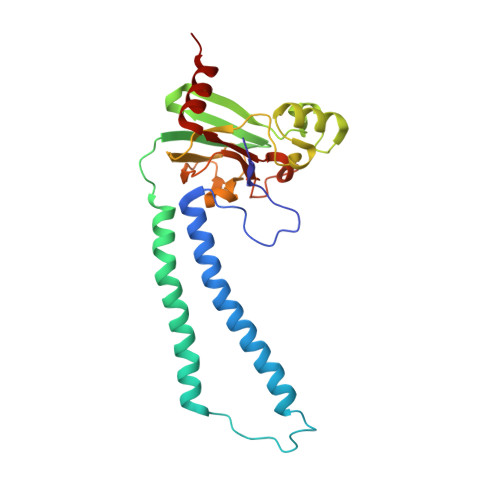
Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Liu, J., Qin, L., Ferguson-Miller, S.(2011) Proc Natl Acad Sci U S A 108: 1284-1289
- PubMed: 21205904
- DOI: https://doi.org/10.1073/pnas.1012846108
- Primary Citation of Related Structures:
3OM3, 3OMA, 3OMI, 3OMN - PubMed Abstract:
Crystal structures in both oxidized and reduced forms are reported for two bacterial cytochrome c oxidase mutants that define the D and K proton paths, showing conformational change in response to reduction and the loss of strategic waters that can account for inhibition of proton transfer. In the oxidized state both mutants of the Rhodobacter sphaeroides enzyme, D132A and K362M, show overall structures similar to wild type, indicating no long-range effects of mutation. In the reduced state, the mutants show an altered conformation similar to that seen in reduced wild type, confirming this reproducible, reversible response to reduction. In the strongly inhibited D132A mutant, positions of residues and waters in the D pathway are unaffected except in the entry region close to the mutation, where a chloride ion replaces the missing carboxyl and a 2-Å shift in N207 results in loss of its associated water. In K362M, the methionine occupies the same position as the original lysine, but K362- and T359-associated waters in the wild-type structure are missing, likely accounting for the severe inhibition. Spectra of oxidized frozen crystals taken during X-ray radiation show metal center reduction, but indicate development of a strained configuration that only relaxes to a native form upon annealing. Resistance of the frozen crystal to structural change clarifies why the oxidized conformation is observable and supports the conclusion that the reduced conformation has functional significance. A mechanism is described that explains the conformational change and the incomplete response of the D-path mutant.
- Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI 48824, USA.
Organizational Affiliation: