Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM.
Chen, J.Z., Settembre, E.C., Aoki, S.T., Zhang, X., Bellamy, A.R., Dormitzer, P.R., Harrison, S.C., Grigorieff, N.(2009) Proc Natl Acad Sci U S A 106: 10644-10648
- PubMed: 19487668
- DOI: https://doi.org/10.1073/pnas.0904024106
- Primary Citation of Related Structures:
3GZT, 3GZU - PubMed Abstract:
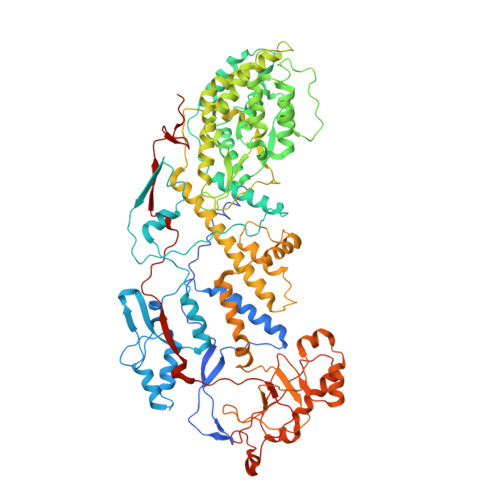
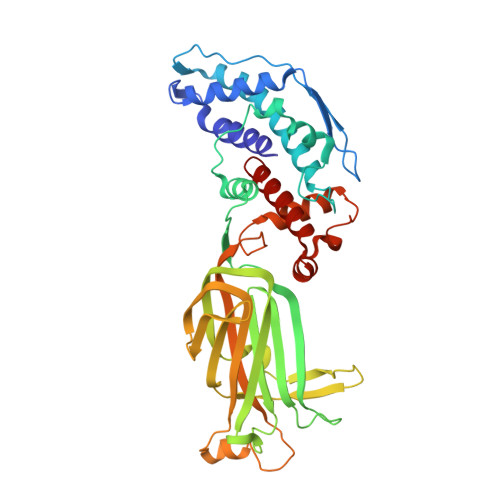
Rotaviruses, major causes of childhood gastroenteritis, are nonenveloped, icosahedral particles with double-strand RNA genomes. By the use of electron cryomicroscopy and single-particle reconstruction, we have visualized a rotavirus particle comprising the inner capsid coated with the trimeric outer-layer protein, VP7, at a resolution (4 A) comparable with that of X-ray crystallography. We have traced the VP7 polypeptide chain, including parts not seen in its X-ray crystal structure. The 3 well-ordered, 30-residue, N-terminal "arms" of each VP7 trimer grip the underlying trimer of VP6, an inner-capsid protein. Structural differences between free and particle-bound VP7 and between free and VP7-coated inner capsids may regulate mRNA transcription and release. The Ca(2+)-stabilized VP7 intratrimer contact region, which presents important neutralizing epitopes, is unaltered upon capsid binding.
- Rosenstiel Basic Medical Research Center, Brandeis University, Waltham, MA 02454, USA.
Organizational Affiliation: