Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens
Singer, A.U., Brown, G., Proudfoot, M., Xu, X., Dong, A., Cui, H., Edwards, A.M., Joachimiak, A., Savchenko, A., Yakunin, A.F.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| uncharacterized protein ATU1752 | 165 | Agrobacterium fabrum str. C58 | Mutation(s): 0 Gene Names: AGR_C_3216, Atu1752 | 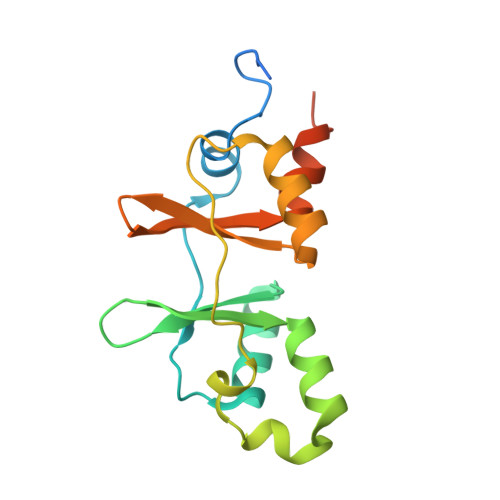 | |
UniProt | |||||
Find proteins for A9CIP4 (Agrobacterium fabrum (strain C58 / ATCC 33970)) Explore A9CIP4 Go to UniProtKB: A9CIP4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A9CIP4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAI Query on NAI | F [auth A], I [auth B], L [auth C], O [auth D] | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE C21 H29 N7 O14 P2 BOPGDPNILDQYTO-NNYOXOHSSA-N |  | ||
| AMP Query on AMP | E [auth A], H [auth B], K [auth C], N [auth D] | ADENOSINE MONOPHOSPHATE C10 H14 N5 O7 P UDMBCSSLTHHNCD-KQYNXXCUSA-N |  | ||
| SO4 Query on SO4 | G [auth A], J [auth B], M [auth C], P [auth D] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 53.247 | α = 115.21 |
| b = 58.075 | β = 106.15 |
| c = 59.056 | γ = 94.94 |
| Software Name | Purpose |
|---|---|
| HKL-3000 | data collection |
| MrBUMP | phasing |
| REFMAC | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |