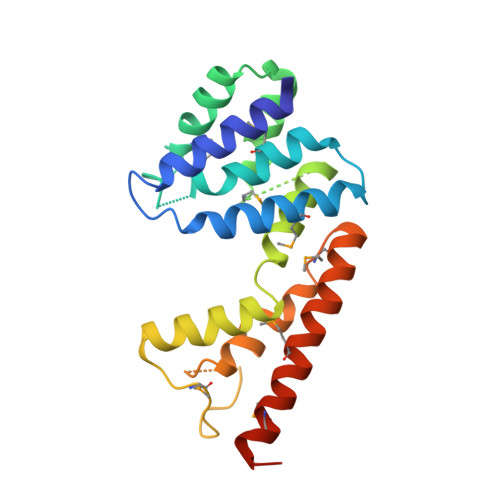
Crystal structure of a HD-superfamily hydrolase (BT9727_1981) from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR114
Forouhar, F., Su, M., Seetharaman, J., Vorobiev, S.M., Janjua, H., Fang, Y., Xiao, R., Cunningham, K., Maglaqui, M., Owen, L.A., Wang, D., Baran, M.C., Liu, J., Acton, T.B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.