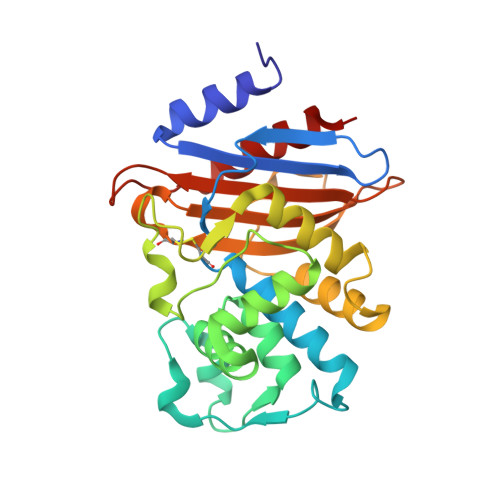
Genetic and structural insights into the dissemination potential of the extremely-broad-spectrum class A {beta}-lactamase (EBSBL) KPC-2 identified in two strains of Escherichia coli and Enterobacter cloacae isolated from the same patient in France
Petrella, S., Ziental-Gelus, N., Mayer, C., Renard, M., Jarlier, V., Sougakoff, W.(2008) Antimicrob Agents Chemother