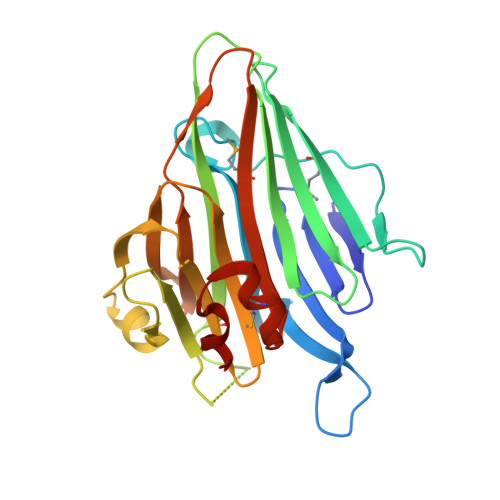
The Crystal Structure of the Periplasmic Domain of the Escherichia coli Membrane Protein Insertase YidC Contains a Substrate Binding Cleft
Ravaud, S., Stjepanovic, G., Wild, K., Sinning, I.(2008) J Biological Chem 283: 9350-9358
- PubMed: 18234665
- DOI: https://doi.org/10.1074/jbc.M710493200
- Primary Citation of Related Structures:
3BS6 - PubMed Abstract:
In bacteria the biogenesis of inner membrane proteins requires targeting and insertion factors such as the signal recognition particle and the Sec translocon. YidC is an essential membrane protein involved in the insertion of inner membrane proteins together with the Sec translocon, but also as a separate entity. YidC of Escherichia coli is a member of the conserved YidC (in bacteria)/Oxa1 (in mitochondria)/Alb3 (in chloroplasts) protein family and contains six transmembrane segments and a large periplasmic domain (P1). We determined the crystal structure of the periplasmic domain of YidC from E. coli (P1D) at 1.8 A resolution. The structure of P1D shows the conserved beta-supersandwich fold of carbohydrate-binding proteins and an alpha-helical linker region at the C terminus that packs against the beta-supersandwich by a highly conserved interface. P1D exhibits an elongated cleft of similar architecture as found in the structural homologs. However, the electrostatic properties and molecular details of the cleft make it unlikely to interact with carbohydrate substrates. The cleft in P1D is occupied by a polyethylene glycol molecule suggesting an elongated peptide or acyl chain as a natural ligand. The region of P1D previously reported to interact with SecF maps to a surface area in the vicinity of the cleft. The conserved C-terminal region of the P1 domain was reported to be essential for the membrane insertase function of YidC. The analysis of this region suggests a role in membrane interaction and/or in the regulation of YidC interaction with binding partners.
- Biochemie-Zentrum der Universität Heidelberg, Im Neuenheimer Feld 328, Heidelberg, Germany.
Organizational Affiliation: