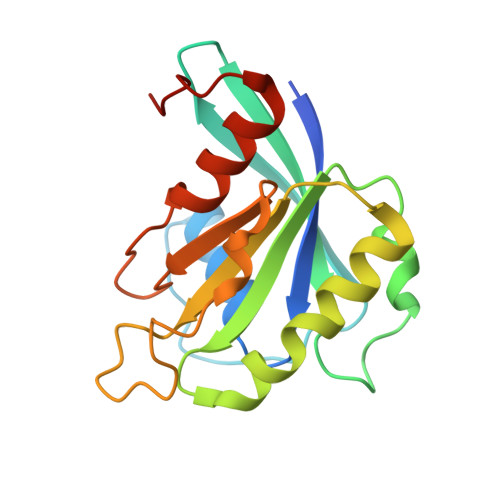
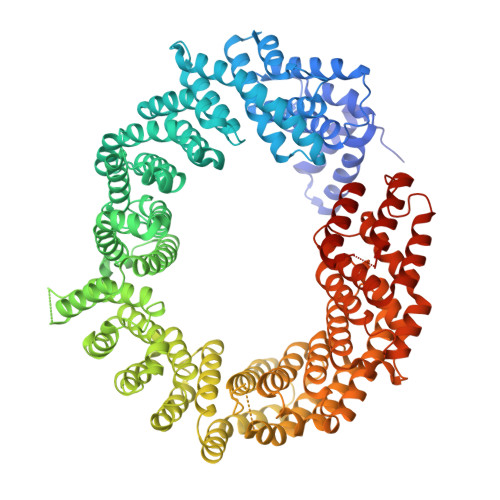
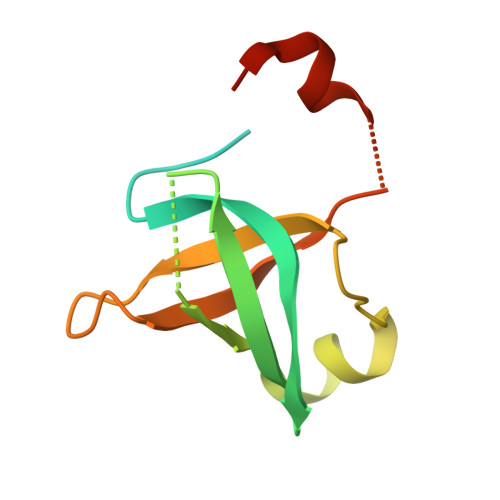
Structural Basis for the Nuclear Export Activity of Importin13.
Grunwald, M., Lazzaretti, D., Bono, F.(2013) EMBO J 32: 899
- PubMed: 23435562
- DOI: https://doi.org/10.1038/emboj.2013.29
- Primary Citation of Related Structures:
3ZJY, 3ZKV - PubMed Abstract:
Importin13 (Imp13) is a bidirectional karyopherin that can mediate both import and export of cargoes. Imp13 recognizes several import cargoes, which include the exon junction complex components Mago-Y14 and the E2 SUMO-conjugating enzyme Ubc9, and one known export cargo, the translation initiation factor 1A (eIF1A). To understand how Imp13 can perform double duty, we determined the 3.6-Å crystal structure of Imp13 in complex with RanGTP and with eIF1A. eIF1A binds at the inner surface of the Imp13 C-terminal arch adjacent and concomitantly to RanGTP illustrating how eIF1A can be exported by Imp13. Moreover, the 3.0-Å structure of Imp13 in its unbound state reveals the existence of an open conformation in the cytoplasm that explains export cargo release and completes the export branch of the Imp13 pathway. Finally, we demonstrate that Imp13 is able to bind and export eIF1A in vivo and that its function is essential.
- Max Planck Institute for Developmental Biology, Tübingen, Germany.
Organizational Affiliation: