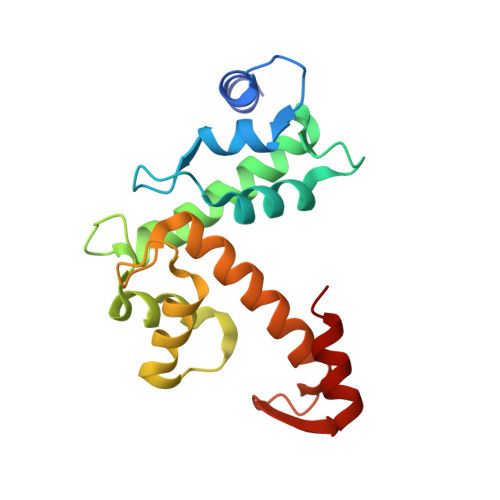
Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2
Takahashi, T., Kojima, K., Zhang, W., Sasaki, K., Ito, M., Suzuki, H., Kawasaki, M., Wakatsuki, S., Takahara, T., Shibata, H., Maki, M.(2015) Int J Mol Sci 16: 3677-3699
- PubMed: 25667979
- DOI: https://doi.org/10.3390/ijms16023677
- Primary Citation of Related Structures:
3WXA - PubMed Abstract:
ALG-2, a 22-kDa penta-EF-hand protein, is involved in cell death, signal transduction, membrane trafficking, etc., by interacting with various proteins in mammalian cells in a Ca2+-dependent manner. Most known ALG-2-interacting proteins contain proline-rich regions in which either PPYPXnYP (type 1 motif) or PXPGF (type 2 motif) is commonly found. Previous X-ray crystal structural analysis of the complex between ALG-2 and an ALIX peptide revealed that the peptide binds to the two hydrophobic pockets. In the present study, we resolved the crystal structure of the complex between ALG-2 and a peptide of Sec31A (outer shell component of coat complex II, COPII; containing the type 2 motif) and found that the peptide binds to the third hydrophobic pocket (Pocket 3). While amino acid substitution of Phe85, a Pocket 3 residue, with Ala abrogated the interaction with Sec31A, it did not affect the interaction with ALIX. On the other hand, amino acid substitution of Tyr180, a Pocket 1 residue, with Ala caused loss of binding to ALIX, but maintained binding to Sec31A. We conclude that ALG-2 recognizes two types of motifs at different hydrophobic surfaces. Furthermore, based on the results of serial mutational analysis of the ALG-2-binding sites in Sec31A, the type 2 motif was newly defined.
- Department of Applied Molecular Biosciences, Graduate School of Bioagricultural Sciences, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8601, Japan. takahashi.takeshi@a.mbox.nagoya-u.ac.jp.
Organizational Affiliation: