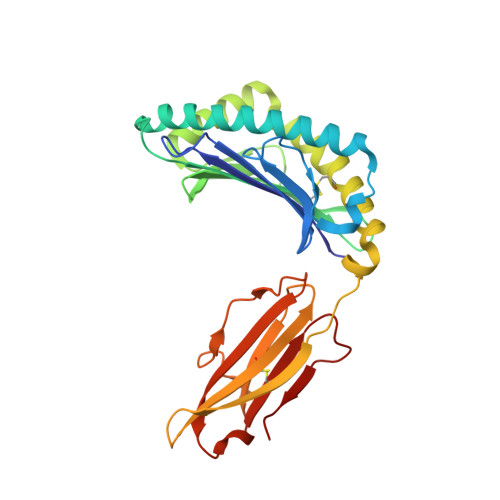
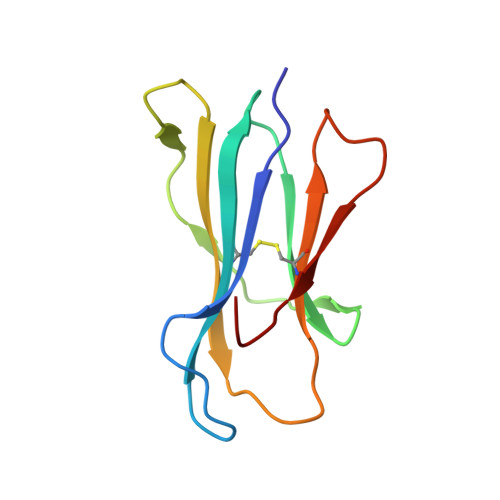
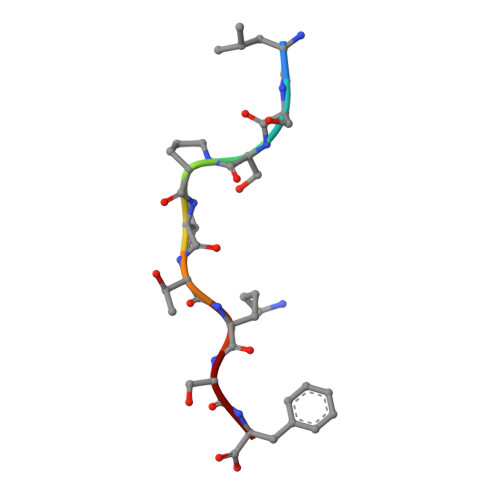
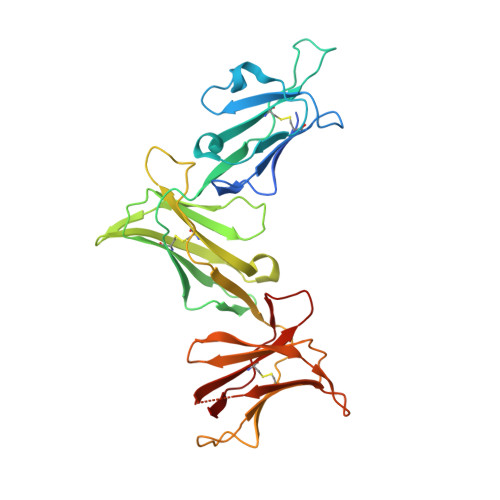
Mutational and structural analysis of KIR3DL1 reveals a lineage-defining allotypic dimorphism that impacts both HLA and peptide sensitivity
O'connor, G.M., Vivian, J.P., Widjaja, J.M., Bridgeman, J.S., Gostick, E., Lafont, B.A.P., Anderson, S.K., Price, D.A., Brooks, A.G., Rossjohn, J., Mcvicar, D.W.(2014) J Immunol 192: 2875-2884
- PubMed: 24563253
- DOI: https://doi.org/10.4049/jimmunol.1303142
- Primary Citation of Related Structures:
3WUW - PubMed Abstract:
Killer Ig-like receptors (KIRs) control the activation of human NK cells via interactions with peptide-laden HLAs. KIR3DL1 is a highly polymorphic inhibitory receptor that recognizes a diverse array of HLA molecules expressing the Bw4 epitope, a group with multiple polymorphisms incorporating variants within the Bw4 motif. Genetic studies suggest that KIR3DL1 variation has functional significance in several disease states, including HIV infection. However, owing to differences across KIR3DL1 allotypes, HLA-Bw4, and associated peptides, the mechanistic link with biological outcome remains unclear. In this study, we elucidated the impact of KIR3DL1 polymorphism on peptide-laden HLA recognition. Mutational analysis revealed that KIR residues involved in water-mediated contacts with the HLA-presented peptide influence peptide binding specificity. In particular, residue 282 (glutamate) in the D2 domain underpins the lack of tolerance of negatively charged C-terminal peptide residues. Allotypic KIR3DL1 variants, defined by neighboring residue 283, displayed differential sensitivities to HLA-bound peptide, including the variable HLA-B*57:01-restricted HIV-1 Gag-derived epitope TW10. Residue 283, which has undergone positive selection during the evolution of human KIRs, also played a central role in Bw4 subtype recognition by KIR3DL1. Collectively, our findings uncover a common molecular regulator that controls HLA and peptide discrimination without participating directly in peptide-laden HLA interactions. Furthermore, they provide insight into the mechanics of interaction and generate simple, easily assessed criteria for the definition of KIR3DL1 functional groupings that will be relevant in many clinical applications, including bone marrow transplantation.
- Cancer and Inflammation Program, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702;
Organizational Affiliation: