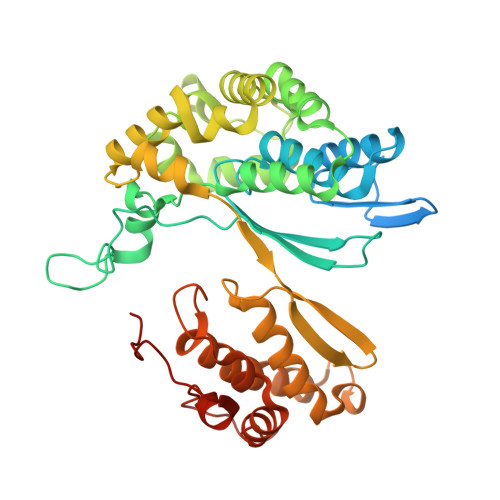
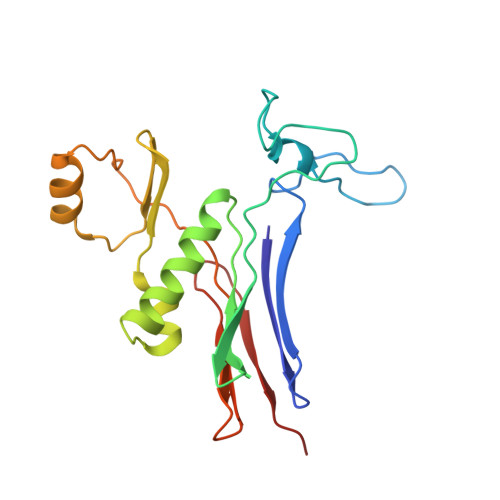
Structure of Bacillus subtilis gamma-glutamyltranspeptidase in complex with acivicin: diversity of the binding mode of a classical and electrophilic active-site-directed glutamate analogue.
Ida, T., Suzuki, H., Fukuyama, K., Hiratake, J., Wada, K.(2014) Acta Crystallogr D Biol Crystallogr 70: 607-614
- PubMed: 24531494
- DOI: https://doi.org/10.1107/S1399004713031222
- Primary Citation of Related Structures:
3WHQ, 3WHR, 3WHS - PubMed Abstract:
γ-Glutamyltranspeptidase (GGT) is an enzyme that plays a central role in glutathione metabolism, and acivicin is a classical inhibitor of GGT. Here, the structure of acivicin bound to Bacillus subtilis GGT determined by X-ray crystallography to 1.8 Å resolution is presented, in which it binds to the active site in a similar manner to that in Helicobacter pylori GGT, but in a different binding mode to that in Escherichia coli GGT. In B. subtilis GGT, acivicin is bound covalently through its C3 atom with sp2 hybridization to Thr403 Oγ, the catalytic nucleophile of the enzyme. The results show that acivicin-binding sites are common, but the binding manners and orientations of its five-membered dihydroisoxazole ring are diverse in the binding pockets of GGTs.
- Department of Biological Sciences, Graduate School of Science, Osaka University, Toyonaka, Osaka 560-0043, Japan.
Organizational Affiliation: