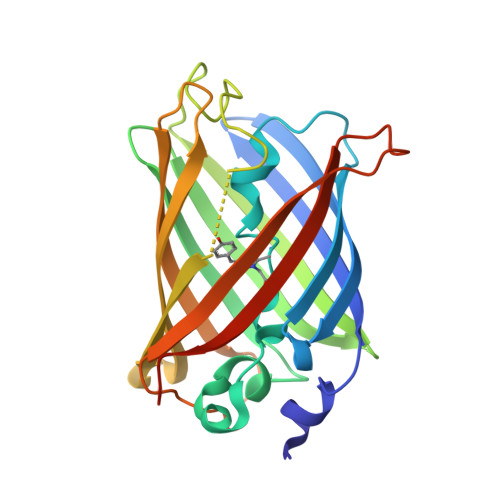
Glycine insertion makes yellow fluorescent protein sensitive to hydrostatic pressure.
Watanabe, T.M., Imada, K., Yoshizawa, K., Nishiyama, M., Kato, C., Abe, F., Morikawa, T.J., Kinoshita, M., Fujita, H., Yanagida, T.(2013) PLoS One 8: e73212-e73212
- PubMed: 24014139
- DOI: https://doi.org/10.1371/journal.pone.0073212
- Primary Citation of Related Structures:
3W1C, 3W1D - PubMed Abstract:
Fluorescent protein-based indicators for intracellular environment conditions such as pH and ion concentrations are commonly used to study the status and dynamics of living cells. Despite being an important factor in many biological processes, the development of an indicator for the physicochemical state of water, such as pressure, viscosity and temperature, however, has been neglected. We here found a novel mutation that dramatically enhances the pressure dependency of the yellow fluorescent protein (YFP) by inserting several glycines into it. The crystal structure of the mutant showed that the tyrosine near the chromophore flipped toward the outside of the β-can structure, resulting in the entry of a few water molecules near the chromophore. In response to changes in hydrostatic pressure, a spectrum shift and an intensity change of the fluorescence were observed. By measuring the fluorescence of the YFP mutant, we succeeded in measuring the intracellular pressure change in living cell. This study shows a new strategy of design to engineer fluorescent protein indicators to sense hydrostatic pressure.
- RIKEN Quantitative Biology Center (QBiC), Suita, Osaka, Japan ; PRESTO, Japan Science and Technology Agency, Kawaguchi, Saitama, Japan ; WPI, Immunology Frontier Research Center, Osaka University, Suita, Osaka, Japan ; Graduate School of Frontier Bioscience, Osaka University, Suita, Osaka, Japan.
Organizational Affiliation: