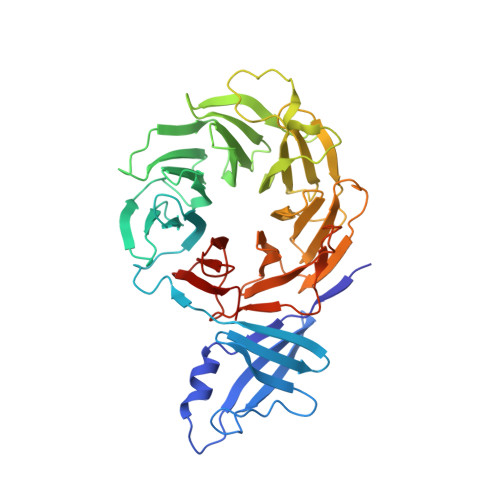
New crystal structure of the proteasome-dedicated chaperone Rpn14 at 1.6 A resolution
Kim, S., Nishide, A., Saeki, Y., Takagi, K., Tanaka, K., Kato, K., Mizushima, T.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 517-521
- PubMed: 22691779
- DOI: https://doi.org/10.1107/S1744309112011359
- Primary Citation of Related Structures:
3VL1 - PubMed Abstract:
The 26S proteasome is an ATP-dependent protease responsible for selective degradation of polyubiquitylated proteins. Recent studies have suggested that proteasome assembly is a highly ordered multi-step process assisted by specific chaperones. Rpn14, an assembly chaperone for ATPase-ring formation, specifically recognizes the ATPase subunit Rpt6. The structure of Rpn14 at 2.0 Å resolution in space group P6(4) has previously been reported, but the detailed mechanism of Rpn14 function remains unclear. Here, a new crystal structure of Rpn14 with an E384A mutation is presented in space group P2(1) at 1.6 Å resolution. This high-resolution structure provides a framework for understanding proteasome assembly.
- Graduate School of Pharmaceutical Sciences, Nagoya City University, 3-1 Tanabe-dori, Mizuho-ku, Nagoya 467-8603, Japan.
Organizational Affiliation: