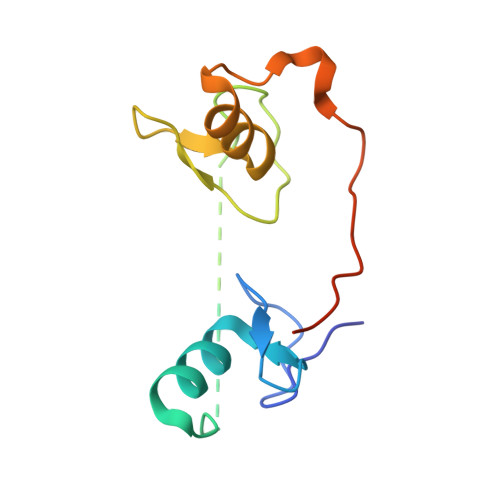
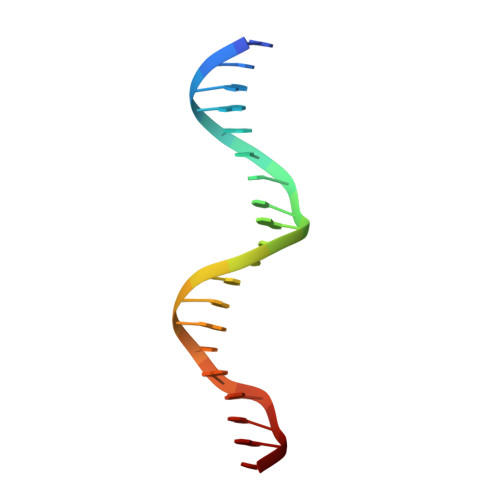
GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Wilkinson-White, L., Lester, K.L., Ripin, N., Jacques, D.A., Mitchell Guss, J., Matthews, J.M.(2015) Protein Sci 24: 1649-1659
- PubMed: 26234528
- DOI: https://doi.org/10.1002/pro.2760
- Primary Citation of Related Structures:
3VD6, 3VEK - PubMed Abstract:
The transcription factor GATA1 helps regulate the expression of thousands of genes involved in blood development, by binding to single or double GATA sites on DNA. An important part of gene activation is chromatin looping, the bringing together of DNA elements that lie up to many thousands of basepairs apart in the genome. It was recently suggested, based on studies of the closely related protein GATA3, that GATA-mediated looping may involve interactions of each of two zinc fingers (ZF) with distantly spaced DNA elements. Here we present a structure of the GATA1 ZF region bound to pseudopalindromic double GATA site DNA, which is structurally equivalent to a recently-solved GATA3-DNA complex. However, extensive analysis of GATA1-DNA binding indicates that although the N-terminal ZF (NF) can modulate GATA1-DNA binding, under physiological conditions the NF binds DNA so poorly that it cannot play a direct role in DNA-looping. Rather, the ability of the NF to stabilize transcriptional complexes through protein-protein interactions, and thereby recruit looping factors such as Ldb1, provides a more compelling model for GATA-mediated looping.
- School of Molecular Bioscience, The University of Sydney, Sydney, New South Wales, 2042, Australia.
Organizational Affiliation: