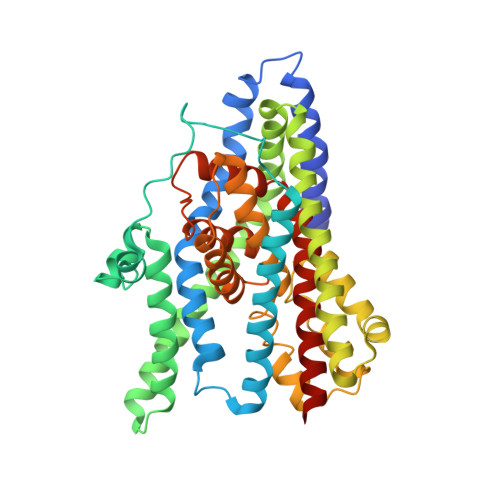
Crystal structure of an asymmetric trimer of a bacterial glutamate transporter homolog.
Verdon, G., Boudker, O.(2012) Nat Struct Mol Biol 19: 355-357
- PubMed: 22343718
- DOI: https://doi.org/10.1038/nsmb.2233
- Primary Citation of Related Structures:
3V8F, 3V8G - PubMed Abstract:
We report a structure of a trimeric glutamate transporter homolog from Pyrococcus horikoshii with two protomers in an inward facing state and the third in an intermediate conformation between the outward and inward facing states. The intermediate shows a cavity in the thinnest region of the transporter, which is potentially accessible to extracellular and cytoplasmic solutions. Our findings suggest a structural principle by which transport intermediates may mediate uncoupled permeation of polar solutes.
- Department of Physiology and Biophysics, Weill Cornell Medical College, New York, New York, USA.
Organizational Affiliation: